M
ethicillin-Resistant Staphylococcus aureus, or MRSA, has been rated by the World Health Organization as the second HIGH priority pathogen in the world, for which new antibiotics need to be discovered. MRSA causes various skin infections/rashes, flesh-eating disease, sepsis, blood infections, pneumonia, and Toxic Shock Syndrome (TSS). In immunocompromised patients, infection with MRSA is especially concerning, and potentially fatal.
"New antibiotics targeting this priority list of pathogens will help to reduce deaths due to resistant infections around the world," says Prof Evelina Tacconelli, Head of the Division of Infectious Diseases at the University of Tübingen and a major contributor to the development of the list. "Waiting any longer will cause further public health problems and dramatically impact on patient care."
MRSA is a gram-positive bacteria resistant to beta-lactam antibiotics. Currently, vancomycin is the most widely used treatment against MRSA infections, however it is an extremely harsh antibiotic; vancomycin therapy has been compared in ferocity of side-effects to chemotherapy. Increasingly, there have been reports of vancomycin failure due to adaptive strains of MRSA becoming resistant to vancomycin.
The most common risk factors for nosocomial acquisition of MRSA are prolonged hospitalization, care in an ICU, prolonged antimicrobial therapy, surgical procedures, and close proximity to a patient in the hospital who is infected/colonized with MRSA. However there has been a rapid increase the incidence of MRSA contracted outside of the hospital. The increase CA-MRSA in non-hospitalized patients is due to the introduction of health care-associated strains into the community. Therefore, effective control of MRSA spread in the community requires effective control of nosocomial MRSA transmission.
The increased spread of MRSA throughout hospitals and into the community is matched with increased concern on how to treat and deal with the bacteria. The regional health authority for Northern British Columbia, Northern Health, indicate the predominate mode of MRSA transmission in healthcare facilities is from one (infected or colonized) patient to another through contaminated hands of healthcare providers. They indicate that those at greatest risk of infection are the elderly, those with chronic diseases and/or those undergoing invasive procedures. Furthermore, statistics from Northern Health indicate that, in comparison to the Antimicrobial Resistance Surveillance Public Health Agency of Canada 2015 infection rates of 0.29 MRSA cases per 1000 pt. days compared to Northern Health infection rates, which were higher at 0.37 per 1000 pt. days in 2015 -2016. This demonstrates the burden MRSA has on Northern communities, a factor heavily propelling our project.
We propose the use of sRNA mediated gene silencing to target specific genes responsible for virulence and drug resistance within MRSA.
W
hy is gene silencing advantageous? The discovery of penicillin in 1928 marked the start of the antibiotic golden age, the following 40 years marked a rapid increase in research and development of new antibiotics. This intense growth saturated the market and lead to our modern, post-antibiotic era. This current era is characterized by concerning rates of antibiotic resistance and a scientific rut, with a 90% decline in the approval of new antibiotics and development of new classes. The scientific rut is perpetuated by a combination of economic, regulatory and scientific barriers. Researchers and physicians alike are looking beyond antibiotics to find a solution to the current antibiotic crisis. Antibiotics, as we have seen, are no longer the solution but a part of the problem as bacteria are able to adapt rapidly to the harsh antibiotics we try to use against them.
We believe this method is superior over antibiotics because it has the capacity to be monitored and altered as needed within hours, whereas antibiotics can take decades to discover.
The nature of DNA evolution is such that mutations must be in sites that will not change the final protein product or regulatory regions, i.e., ribosome binding sites, promoters, and 5’ UTRs. If a sequence does mutate and is selected in the population, our method lends rapid adaptability to evolutionary pressures. When the treatment stops working, the gene of interest can be sequenced, and based on that sequence, the sRNA can be modified accordingly. Furthermore, sRNA mediated gene silencing has shown very promising results in the literature, with knockdown results approximating 99% (Park et al., 2012).
Project Design
Our project has been divided into a two-year plan. This year encompassed identification of vital pathways for MRSA survival, including resistance and virulence, as well what is necessary for sRNA silencing in Staphylococcus species. We identified the following genes as targets for gene silencing via sRNA:
- secA, a critical ATPase protein in the bacterial secretion system. This system is responsible for secreting toxins and glycoproteins responsible for virulence, as well as membrane proteins essential in the organisms survival.
- mecA, a genetic element exclusive to MRSA, which gives the organism resistance to beta-lactams through multiple pathways. This element includes PBP2A, or penicillin binding protein, which binds and inhibits beta-lactams.
- Ddl, short for d-alanine—d-alanine ligase, catalyzes the formation of d-alanine cross linkages within peptidoglycan in the cell wall.
An important consideration made when identifying the above genes is that there are no so-called “back-up” pathways or genes that can take over the function of the gene of interest, should it be shut down fully. After finding the genes of interest, we then created sequences directly complimentary to the mRNA we wished to target, and had our sRNAs synthesized and 5’ FAM labelled by IDT for measurement purposes.
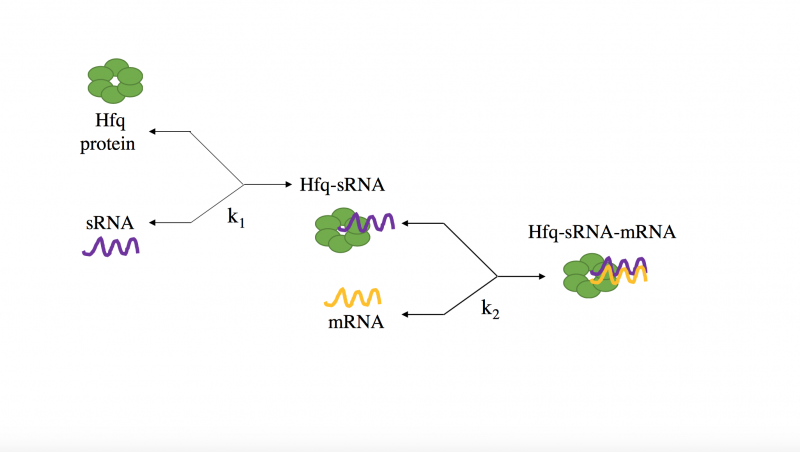
Literature review of dsRNA binding protein showed Hfq to be essential in sRNA mediated gene silencing in E. coli, however there is a lack of general consensus on whether or not it is functional in Staphylococcus species. Therefore, determining if Hfq is needed to stabilize the interaction between sRNA and mRNA in Staphylococcus species became the focus this year’s project; as this is vital information needed to proceed to engineer a successful silencing system.
If Hfq binds to the sRNA, we would know if it is essential to include on the plasmid utilized for our proposed treatment. This plasmid would then not only include the gene that codes for the appropriate sRNA, but would also need to include the gene for Hfq under a bidirectional tetracycline inducible promoter. It will become the focus of next year to engineer and deliver such a plasmid.
Literature on Staphylococcus Hfq demonstrates that it binds most favorably to an AUAUAUA portion of RNA (Horstmann et al., 2012). Based on this, we included an AUAUAUA sequence on the 5' end of our custom designed sRNAs to allow for Hfq binding.
We further developed a qPCR assay for future use in determining the level of gene knockdown in vivo.To test the efficacy of gene silencing in vivo, we designed a qPCR assay around the target genes of interest, Hfq, and two reporter genes: 6-phosphofructokinase (6-PFK) and acetyl-CoA carboxylase (ACC). These reporters were chosen as they are part of crucial metabolic function, and expression levels are expected to be constant, even if gene silencing is occurring at other sites. Furthermore, these genes are genetically distant from the genes of interest, and thus have no probability of being expressed on a polycistronic mRNA.