Zsy5691750 (Talk | contribs) |
|||
| Line 257: | Line 257: | ||
<div class="small_pic_demo" style="float:left;"> | <div class="small_pic_demo" style="float:left;"> | ||
<a href="#pic_ten"> | <a href="#pic_ten"> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/1/1b/Tianjin-ho-result-10.jpeg"></a><p style="font-size:15px;text-align:center"><br/> Fig 1-1 The PCR strategy for testing whether we deleted the HMRa in <b><i>SynⅩ-dUra</i></b>. | + | <img src="https://static.igem.org/mediawiki/2017/1/1b/Tianjin-ho-result-10.jpeg"></a><p style="font-size:15px;text-align:center"><br/> Fig. 1-1. The PCR strategy for testing whether we deleted the HMRa in <b><i>SynⅩ-dUra</i></b>. |
</p> | </p> | ||
</div> | </div> | ||
| Line 263: | Line 263: | ||
<a href="#pic_eleven" > | <a href="#pic_eleven" > | ||
<img src="https://static.igem.org/mediawiki/2017/0/03/Tianjin-ho-result-9.jpeg"/> | <img src="https://static.igem.org/mediawiki/2017/0/03/Tianjin-ho-result-9.jpeg"/> | ||
| − | </a> <p style="font-size:15px;text-align:center"><br/>Fig 1-2 As we can see in the gel photo above, the <b>UP</b> and <b>DOWN</b> segments hasn’t been amplified in our <i><b>SynⅩ-dUra</b></i> comparing to the BY4741 as control. Which indicated that the HMRa gene has been successfully eliminated. | + | </a> <p style="font-size:15px;text-align:center"><br/>Fig. 1-2. As we can see in the gel photo above, the <b>UP</b> and <b>DOWN</b> segments hasn’t been amplified in our <i><b>SynⅩ-dUra</b></i> comparing to the BY4741 as control. Which indicated that the HMRa gene has been successfully eliminated. |
</p> | </p> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="pic_ten" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/1/1b/Tianjin-ho-result-10.jpeg"/><p style="font-size:15px;text-align:center"><br/> Fig 1-1 The PCR strategy for testing whether we deleted the HMRa in <b><i>SynⅩ-dUra</i></b>. | + | <div id="pic_ten" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/1/1b/Tianjin-ho-result-10.jpeg"/><p style="font-size:15px;text-align:center"><br/> Fig. 1-1. The PCR strategy for testing whether we deleted the HMRa in <b><i>SynⅩ-dUra</i></b>. |
</p></div> | </p></div> | ||
| − | <div id="pic_eleven" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/0/03/Tianjin-ho-result-9.jpeg"/><p style="font-size:15px;text-align:center"><br/>Fig 1-2 As we can see in the gel photo above, the <b>UP</b> and <b>DOWN</b> segments hasn’t been amplified in our <i><b>SynⅩ-dUra</b></i> comparing to the BY4741 as control. Which indicated that the HMRa gene has been successfully eliminated. | + | <div id="pic_eleven" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/0/03/Tianjin-ho-result-9.jpeg"/><p style="font-size:15px;text-align:center"><br/>Fig. 1-2. As we can see in the gel photo above, the <b>UP</b> and <b>DOWN</b> segments hasn’t been amplified in our <i><b>SynⅩ-dUra</b></i> comparing to the BY4741 as control. Which indicated that the HMRa gene has been successfully eliminated. |
</p></div> | </p></div> | ||
<h4> The result for constructing the <i>Gal</i> systems</h4> | <h4> The result for constructing the <i>Gal</i> systems</h4> | ||
| Line 290: | Line 290: | ||
<a href="#pic_twentyone"> | <a href="#pic_twentyone"> | ||
<img src="https://static.igem.org/mediawiki/2017/b/b9/Tianjin-ho-result-666.jpeg"></a> | <img src="https://static.igem.org/mediawiki/2017/b/b9/Tianjin-ho-result-666.jpeg"></a> | ||
| − | <p style="font-size:15px;text-align:center"><br/>Fig 1-3 The results of PCR of #6, #7, #16, #20, #27, #36, #37, #55 colonies. <i>HO</i> gene (length of 1770bp). As we can see, <i>HO</i> gene in all 8 colonies has been amplified, which indicated that we succeeded in constructing the device for <i>HO</i> gene expression.</p> | + | <p style="font-size:15px;text-align:center"><br/>Fig. 1-3. The results of PCR of #6, #7, #16, #20, #27, #36, #37, #55 colonies. <i>HO</i> gene (length of 1770bp). As we can see, <i>HO</i> gene in all 8 colonies has been amplified, which indicated that we succeeded in constructing the device for <i>HO</i> gene expression.</p> |
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="pic_twentyone" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/b/b9/Tianjin-ho-result-666.jpeg"><p style="font-size:15px;text-align:center"><br/>Fig 1-3 The results of PCR of #6, #7, #16, #20, #27, #36, #37, #55 colonies. <i>HO</i> gene (length of 1770bp). As we can see, <i>HO</i> gene in all 8 colonies has been amplified, which indicated that we succeeded in constructing the device for <i>HO</i> gene expression.</p></div> | + | <div id="pic_twentyone" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/b/b9/Tianjin-ho-result-666.jpeg"><p style="font-size:15px;text-align:center"><br/>Fig. 1-3. The results of PCR of #6, #7, #16, #20, #27, #36, #37, #55 colonies. <i>HO</i> gene (length of 1770bp). As we can see, <i>HO</i> gene in all 8 colonies has been amplified, which indicated that we succeeded in constructing the device for <i>HO</i> gene expression.</p></div> |
<h4>The result of mating type switching(<b>MTS</b>)</h4> | <h4>The result of mating type switching(<b>MTS</b>)</h4> | ||
<hr> | <hr> | ||
| Line 321: | Line 321: | ||
<div class="small_pic_demo" style="float:left;"> | <div class="small_pic_demo" style="float:left;"> | ||
<a href="#pic_twentytwo"> | <a href="#pic_twentytwo"> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/7/7e/Tianjin-ho-resultN1.jpeg"></a><p style="font-size:15px;text-align:center"><br/>Fig 1-4 (a) showed the PCR results for MATa locus. The MATa gene was amplified in all colonies except the first 24 colonies. | + | <img src="https://static.igem.org/mediawiki/2017/7/7e/Tianjin-ho-resultN1.jpeg"></a><p style="font-size:15px;text-align:center"><br/>Fig. 1-4. (a) showed the PCR results for MATa locus. The MATa gene was amplified in all colonies except the first 24 colonies. |
</p> | </p> | ||
</div> | </div> | ||
| Line 327: | Line 327: | ||
<a href="#pic_twentythree" > | <a href="#pic_twentythree" > | ||
<img src="https://static.igem.org/mediawiki/2017/b/b7/Tianjin-ho-result-7.jpeg"/> | <img src="https://static.igem.org/mediawiki/2017/b/b7/Tianjin-ho-result-7.jpeg"/> | ||
| − | </a> <p style="font-size:15px;text-align:center"><br/>Fig 1-4 (b) showed the PCR results for MATα locus. The MATα gene was amplified in all 96 colonies.</p> | + | </a> <p style="font-size:15px;text-align:center"><br/>Fig. 1-4. (b) showed the PCR results for MATα locus. The MATα gene was amplified in all 96 colonies.</p> |
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="pic_twentytwo" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/7/7e/Tianjin-ho-resultN1.jpeg"/><p style="font-size:15px;text-align:center"><br/>Fig 1-4 (a) showed the PCR results for MATa locus. The MATa gene was amplified in all colonies except the first 24 colonies. | + | <div id="pic_twentytwo" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/7/7e/Tianjin-ho-resultN1.jpeg"/><p style="font-size:15px;text-align:center"><br/>Fig. 1-4. (a) showed the PCR results for MATa locus. The MATa gene was amplified in all colonies except the first 24 colonies. |
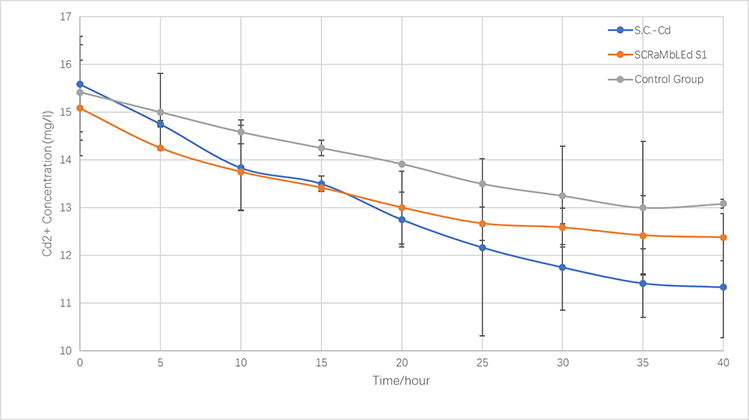
Cadmium concentration: 5mM for the first and the third experiments; 10 mM for the second experiment. | Cadmium concentration: 5mM for the first and the third experiments; 10 mM for the second experiment. | ||
</p></div> | </p></div> | ||
| − | <div id="pic_twentythree" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/b/b7/Tianjin-ho-result-7.jpeg"/><p style="font-size:15px;text-align:center"><br/>Fig 1-4 (b) showed the PCR results for MATα locus. The MATα gene was amplified in all 96 colonies.</p></div> | + | <div id="pic_twentythree" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/b/b7/Tianjin-ho-result-7.jpeg"/><p style="font-size:15px;text-align:center"><br/>Fig. 1-4. (b) showed the PCR results for MATα locus. The MATα gene was amplified in all 96 colonies.</p></div> |
<h4>Discussion & Expectation</h4> | <h4>Discussion & Expectation</h4> | ||
Revision as of 08:12, 30 October 2017
/* OVERRIDE IGEM SETTINGS */