| Line 5,237: | Line 5,237: | ||
</button> | </button> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/ | + | <img src="https://static.igem.org/mediawiki/2017/6/6a/Mingdaophil1026-7.png" width="60%" style="padding-left: 2.5rem"> |
| − | + | ||
<h4 style="padding-left: 5rem"> In the assay for repressor system, the data in Fig. 4 gave the similar results as team Glasgow did in 2015, in which the strong activity of the repressible promoter was significantly repressed in the presence of PhlF repressor.</h4> | <h4 style="padding-left: 5rem"> In the assay for repressor system, the data in Fig. 4 gave the similar results as team Glasgow did in 2015, in which the strong activity of the repressible promoter was significantly repressed in the presence of PhlF repressor.</h4> | ||
Revision as of 00:42, 26 October 2017
INTRODUCTION
INTRODUCTION

Obesity and type 2 diabetes represent a serious health threat to the population all over the world. Glucose overdose has the major impact on this problem. However, sugar does make you feel happy. Some people may choose to limit, or even quit, sugar intake. Others may do more exercise to stay fit and healthy. In plain words, you have to either decline sweet happiness or pay for it with gaining fat or spending hard time burning calories.
In Mingdao’s project this year, we will help you “crush” the sugar by doing nothing except taking one pill full of our engineered probiotics. Really, just sit and read our iGEM story as well as enjoy your sweets and sugary drinks.
In the research, we addressed the health problems caused by excess glucose, providing information of controlling blood glucose by medicine, briefing previous iGEM team projects dealing with the issue of obesity and diabetes, and detail the mechanism of glucose metabolism and pleasure.
In the design and experiment, we will show you how we created probiotic-based glucose retrieval system with (1) glucose transporter device, which facilitates the cell glucose uptake with high efficiency, (2) glucose responsive suicide circuit, which produces lysis protein and nuclease to destroy bacteria when running out of glucose, and (3) probiotic transgenesis system, which be utilized to stably transform Lactobacillus acidophilus by chromosomal homologous recombination.
In the end, we introduced the modeling of the glucose uptake efficiency of our genetically engineered bacteria based on our experimental results. In addition, we showed the design of our acid-resistant pill and the prototype to mimic the stomach-gut environment and simulate the pill passing through gastric acid and releasing probiotics in the gut.

RESEARCH

What does glucose do to your body?
SUGAR exists in the types of glucose, fructose, galactose, sucrose, lactose, maltose and starch, as well as in wide varieties of drinks and food such as juice, honey, fruit, yogurt, candies and desserts. Simply speaking, sugar is everywhere, anyone can’t avoid it. Glucose is the smallest form of sugar and used as energy source for the body. And further, glucose would make you feel pleasure when hitting your tongue letting you crave more sugar like addicted.
OBESITY raises increasing concerns worldwide. Too much sugar in the body will be converted to fat and stored in the liver and cells. Excess sugar will make you overweight and obese. Meanwhile, fat is easily gained but hard to break down.
TYPE 2 DIABETES is another health problem because of excess sugar in the blood for a long term period. Insulin produced by the pancreas stimulates cells to absorb extracellular glucose. But in patients with type 2 diabetes, cells somehow become insulin resistant and need more amounts of insulin to maintain normal function. So the patients are advised to adjust their diet and control the blood sugar level.
The fat converted from excess glucose may contribute to atherosclerosis and increase the risk of HEART DISEASE. In addition, a study provided evidence that high blood glucose could lead to ALZHEIMER'S DISEASE and may damage your brain to cause dementia (Scientific Reports 2016;6:25589).
PROBLEM - Excess Sugar Consumption
How to deal with the excess glucose problems
To stay fit and keep healthy, you can choose to control desire and avoid sugary food or drinks. Or what else you can do to avoid the excess glucose in the blood?
DOING EXERCISE is good to your health and burning glucose you gained. Accordingly, you have to running (at the speed of 5 mph) for 13 min to burn off calories from 330ml of sugary soft drink, 28 min for a medium cup of mocha coffee, 43 min for a sandwich with chicken & bacon or a ¼ pizza (published by Royal Society of Public Health, 2016). When you think about it, I guess you probably already give up.
ARTIFICIAL SWEETENERS in diet drinks are common synthetic substitutes for glucose such as aspartame and saccharine. One survey by Boston University School of Medicine found that diet drinks are associated with higher risk of dementia caused by strokes (published on Stroke, 2017). Another study published on Canadian Medical Association Journal (CMAJ., 2017) reported that artificial sweeteners linked to risk of weight gain and heart disease. The sweeteners may have negative effects on metabolism, gut bacteria and appetite.
TAKING MEDICINE to reduce blood glucose is one of the treatments for diabetes. Canagliflozin is a drug of an inhibitor of subtype 2 sodium-glucose transport proteins (SGLT2) (Can J Diabetes., 2017). SGLT-2 play a major role in renal glucose reabsorption. When blocking SGLT-2 by canagliflozin, the blood glucose could be eliminated through the urine. However, the drug has adverse side effects such as fungal infection, thirst, increased urination and low blood pressure. DPP-4 inhibitor is a relatively new antidiabetic drug (Postgrad Med., 2017). DPP-4 is an enzyme destroying a gastrointestinal hormones called incretins which stimulates insulin production and inhibits the gluconeogenesis by liver. Blocking DPP-4 function by the inhibitor may reduce blood glucose level and lose weight. But DPP-4 inhibitor has severe side effects of nausea, diarrhea and stomach pain, etc.
Exercise to burn off the calories you take from food is not realistic. Artificial sweeteners and diet drinks are considered bad to your health and have higher risk to some diseases. Medicine is the bottom line and you never want to crave food followed by taking pills with side effects. Then, what should you do to deal with the excess glucose?
What have iGEMers done to solve the obesity and diabetes problems?
Problem-solving iGEMers
What have iGEMers done to solve the obesity and diabetes problems?
iGEMers are always tackling real problems happening in the world and seeking a potential solution based on synthetic biology. In the issues of obesity and diabetes, iGEM teams NTU-LIHPAO-Taiwan and Stony_Brook in 2015 contributed to the work of body weight control and diabetes treatment, respectively.
NTU-LIHPAO-Taiwan has developed Appetite Controller to reduce appetite so as to control body weight. They engineered a probiotic, Lactobacillus casei, to produce CPP-PYY fusion polypeptides. Peptide YY (PYY) is one of the gastrointestinal hormones which controls appetite. The PYY is carried by Cell Penetrating Peptide (CPP) to penetrate into the intestinal cells. The products would keep the body fit by lowering appetite when people crave foods but aren’t hungry.
Stony_Brook has engineered a QSP tripeptide secreting E. coli to regulating blood glucose level. The QSP tripeptide is encoded by RSC1A1 gene and acts on kidney cells to inhibit the expression of a glucose transporter (SGLT2) on the cell membrane which facilitates the glucose retrieval. Therefore, the product acts like anti-diabetic drug canagliflozin and would help people with high blood glucose excrete the glucose via urination.
However, if you take NTU-LIHPAO-Taiwan’s product, you may no longer enjoy delicious food. And if you need Stony_Brook’s solution, you’re probably getting some troubles with high blood glucose. Is there any better way to control glucose and enjoy the sweets?
Think about why sweets make you happy
Have you ever thought about why sweets make you feel pleasure?
First, the sugar enters your mouth. When sugar hits your tongue, the carbohydrate molecules are bound to the sweet receptors of cells in taste buds. Then the signal will be transduced to taste center in the brain via taste nerve. And the dopamine reward system turns on and sends pleasure signal to make you feel happiness. Dopamine is a neurotransmitter in brain neuron system that regulates how you perceive and experience pleasure. In addition to sugary foods, when doing enjoyable activities, dopamine is being released and make you feel happy.
Next, the glucose goes to digestive system through esophagus via stomach to the small intestine. Glucose in there will be absorbed into intestinal cells by cell membrane Sodium Glucose Co-transporter-1 (SGLT1) and go out to bloodstream into the circulatory system by Glucose transporter 2 (GLUT2).
Then, blood glucose stimulates the pancreas to release insulin which enhances glucose entry into the cells and lower the blood glucose level. However, when blood glucose level is out of control for a long time, the cells may become insulin-resistant and that’s so-called type 2 diabetes.
Finally, the excess glucose will be converted to glycogen in liver and muscle, as well as stored as fat in adipose tissue. Extra fat will let you gain weight and eventually be obese.
What will MINGDAO iGEM team do?
Take a look back and think for a while, in which way you can enjoy sweet happiness and not increase the risk of obesity and diabetes. YES! You got an idea! Block the glucose absorption from entry into the small intestine. You highjack the sugar. Next, we will show you how we “crush” sugar in iGEM project this year.
- Reference -
- 1. Type 3 Diabetes: Cross Talk between Differentially Regulated Proteins of Type 2 Diabetes Mellitus and Alzheimer's Disease. Sci Rep. 2016;6:25589
- 2. Ten calorie dense food and their activity equivalence. Royal Society for Public Health Publication, 2016.
- 3. Sugar- and Artificially Sweetened Beverages and the Risks of Incident Stroke and Dementia: A Prospective Cohort Study. Stroke. 2017;48(5):1139-1146.
- 4. Nonnutritive sweeteners and cardiometabolic health: a systematic review and meta-analysis of randomized controlled trials and prospective cohort studies. CMAJ. 2017;189(28):E929-E939.
- 5. The Role of Sodium-Glucose Cotransporter 2 Inhibitors in the Management of Type 2 Diabetes. Can J Diabetes. 2017;41(5):517-523.
- 6. SGLT2 inhibitor/DPP-4 inhibitor combination therapy - complementary mechanisms of action for management of type 2 diabetes mellitus. Postgrad Med. 2017;129(4):409-420.
DESIGN

- BioBrick Blueprint & Prototype Design -
CP29-RBS-aeBlue-RBS-STM1128-TT/pSB1C3 (BBa_K2230028)
A facilitated sodium/glucose cotransporter encoded by STM1128 was cloned out from Salmonella typhimurium LT2. Promoter CP29 is a strong and constitutive promoter working in both E. coli and Lactobacillus spp. The device also displays blue color to demonstrate the engineered bacteria.
Pcar-wRBS-PhlF-T-Pr-sRBS-GFP-sRBS-lysis-sRBS-NucA/pSB1C3 (BBa_K2230017)
The device includes a glucose responsive promoter (Pcar), a repressor circuit (PhlF & its repressed promoter), and a suicide switch composed of lysis protein (lysis) and nuclease (NucA) to destroy cell membrane and chromosomal DNAs. In the presence of glucose, the repressor PhlF is expressed and inhibit the corresponding promoter. When glucose runs out, the PhlF is gradually degraded and the suicide circuit will then turn on to kill the host. The device also carries GFP for detection and measurement.
RBS-EmR-CP29-RBS-aeBlue/pLBA169 (BBa_K2230004)
pLBA169 was designed as vector for transforming Lactobacillus acidophilus thru chromosome homologous recombination. The gene cassette will be inserted to the location at the downstream of slpA (LBA0169) which encodes a surface layer protein. EmR (erythromycin resistance gene) is driven by the upstream promoter of slpA and acts as a selection marker. Promoter CP29 drives gene expression of aeBlue in Lactobacillus.
Our products will be encapsulated into the pill. Microencapsulation process combines sodium alginate and calcium chloride to form microspheres. This tiny spheres are acid resistant and don’t release probiotics in gastric fluid until passing into higher pH (pH=7~9) in the environment of the intestinal tract.

DEMONSTRATION

Glucose is transported into the small intestine and from there into the blood. Na+-glucose cotransporter SGLT1 is involved in intestinal sugar absorption, and Glucose transporter 2 (GLUT2) facilitate glucose transportation from intestine to blood stream.


Which glucose transporter should we use?
In order to compete the absorption of glucose with intestine, we need a better transporter for glucose uptake with higher efficiency. The value of Km of an enzyme was utilized as a parameter to select our target. Km (Michaelis constant) is determined by the concentration of substrate which permits the enzyme to achieve half Vmax. The lower Km means a higher substrate affinity.
As you can see in the right table, based on our research, the glucose transporter of Salmonella typhimurium has lowest Km, meaning the highest affinity of glucose bound to the transporter. So we decided to choose glucose transporter system of Salmonella as our target.

FACT
Interestingly, Salmonella is an intracellular intestinal pathogen. To survive in the small intestine epithelial cells, Salmonella has to utilize and metabolize available and limited glucose in a multiple and efficient way. Not surprisingly, Salmonella has higher affinity of glucose transporter compared to human small intestine. And it all makes sense to our assumption.
- - REFERENCE -
- 1. Glucose Galactose Malabsorption. American Journal of Physiology - Gastrointestinal and Liver Physiology 1998;275:G879-G882
- 2. Functional Properties and Genomics of Glucose Transporters. Curr Genomics. 2007;8(2): 113–128.
- 3. The SLC2 (GLUT) Family of Membrane Transporters. Mol Aspects Med. 2013;34(0): 121–138.
- 4. Glucose and Glycolysis Are Required for the Successful Infection of Macrophages and Mice by Salmonella enterica Serovar Typhimurium. Infect Immun. 2009;77(7): 3117–3126.
Gene Cloning
Salmonella typhimurium LT2 has two glucose-specific transporter systems, PTS system and sodium/glucose cotransporter. PTS system contains two subunits IIA encoded by crr and IIBC by ptsG which are assembled to a high-affinity active transporter. The other is a Na+/glucose cotransporter encoded by STM1128 that contributes to facilitated transport with lower glucose affinity. We decided to genetically engineer microbes with these two systems.
In order to express the genes in E. coli for demonstration and in probiotics for proof-of-concept in a real world. We chose promoter CP29 that is a strong constitutive promoter working well in both E. coli and Lactobacillus spp1. The biobrick part, CP29-RBS-aeBlue (BBa_K1033280) was used and to be assembled with the transporter genes.

Problem 1.
– Optimized DNA sequences can’t be synthesized
Integrated DNA Technologies, Inc. (IDT) contributes to synthetic biology community and is kindly providing free DNA synthesis service for iGEM teams every year. We first designed the genes by optimizing the gene sequence for expression in both E. coli and Lactobacillus spp. and expected to acquire gene synthesis from IDT.
Three weeks passed, IDT has tried hard but not made the synthetic gene successfully. In the end, we knew that it’s probably because the overexpression of glucose transporter genes are somehow toxic to E. coli. Therefore, IDT can’t synthesize the genes which are linked to a strong promoter.
Trouble-shooting 1.
– Amplification of genes by PCR with gDNA
We found a way to directly clone the genes out from Salmonella. Unfortunately, Salmonella is a human pathogen and classify as Biosafety Level 2 agents. First of all, we check the virulence factors (e.g., invasion/adhesion proteins, fimbriae, flagella, type I and III secretion systems)2 and confirm the glucose transporter genes are out of the list.
We sought collaboration with other iGEM teams to got the genes. Luckily, the place of iGEM team CSMU_NCHU _Taiwan is just near our school campus and working in a lab with the materials of Salmonella. They helped us amplify the genes of crr, ptsG and STM1128 by PCR using genomic DNA of Salmonella typhimurium LT2 as template (the gDNAs are recognized as Biosafety Level 1 agents, according the information provided by ATCC).

Now we could move forward with the PCR-amplified DNA fragments of glucose transporter genes of Salmonella.
Problem 2.
– Promoter was gone in the recombinant DNA
We soon assembled RBS-crr and RBS-ptsG to Double terminator/pSB1C3 (BBa_B0015) to make RBS-crr-RBS-pstG-TT/pSB1C3, so did to RBS-STM1128 to make RBS-STM1128-TT/pSB1C3. But something unexpectedly was happening in the next step.
We are unable to assemble both of the transporter composite parts with Double terminator under CP29 promoter.
A paper demonstrated growth inhibition by elevated transport of sugar phosphates in E. coli with overexpression of a glucose transporter (uhpT) gene3. Sugar phosphates transported through the transporter directly enter into glycolytic pathway resulting in the accumulation of toxic metabolite, methylglyoxal which causes the bacterial growth inhibition or cell death. It could also occur in our situation.
Although different and regulated promoters like PBAD or Plac could save our gene cloning, we still continued to focus on the CP promoter because a feasible promoter working both on E. coli and Lactobacilus spp. and easily applied to industrial manufacture without addition of any chemicals (inducers) are what we want.
Trouble-shooting 2.
– Different culture media replacing LB
Based on the fact that excess glucose entering bacterial cell at a time stresses the survival of E. coli, we were working in other strategies or with substitutes as energy source.
First, we tried to use agar plates made of M9 minimal salt media with a series of reducing concentration of glucose (i.e, 20mM, 10mM, 5mM, 2.5mM, 1.25mM, 0mM). The results showed that the colonies formed are either carrying CP29 promoter only or containing RBS and transporter genes without any promoters.
Secondly, we learned that MacConkey broth replace glucose with lactose for fermentation studies. We transformed E. coli with recombinant DNAs and grew them on MacConkey agar plate. We screened dozens of colonies by PCR and luckily found one containing CP29-RBS-aeBlue-RBS-crr-RBS-ptsG/pSB1C3. The plasmids were extracted and checked with restriction enzyme, as well as further confirmed by sequencing.

Finally, we thought another carbon sources in addition to glucose and lactose. This time, M9 media with glycerol were used. Luckily again, one colony carrying CP29-RBS-aeBlue-RBS-STM1128 was picked up by us. And we’ve check it with PCR, restriction enzyme and sequencing.

For more gene cloning detail, please go to our PARTS section.
- - REFERENCE -
- 1. The Sequence of Spacers between the Consensus Sequences Modulates the Strength of Prokaryotic Promoters. Appl Environ Microbiol. 1998;64(1): 82–87.
- 2. Salmonella – the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11(11): 1579–1586.
- 3. Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J Gen Microbiol. 1992;138(10):2007-14.
Demonstration
Experiment





To measure glucose uptake by the engineered E. coli expressing PTS system or Na+/glucose cotransporter, the bacteria were culture in LB broth supplemented with 34μg/ml of chloramphenicol at 37°C overnight. The next day, the bacterial culture was adjusted to OD600 = 3 and exchanged with M9 minimal media with 20mM of glucose for 4 hours or at different time points.


Glucose concentration was analyzed with Glucose (HK) Assay Kit (Sigma-Aldrich) according to the manufacturer’s instruction. Briefly, glucose was phosphorylated (G6P) by hexokinase. Then G6P was further catalyzed by G6PDH and the reduced NAHD was formed from the oxidation of NAD, resulting in increasing in absorbance at 340 nm.

Result
As shown in Fig. 3, the growth of E. coli expressing the PTS reporter (i.e., crr and ptsG genes) was seriously retardant. As I mentioned earlier, the E. coli overexpressing glucose transporter may lose viability because of the toxic metabolites produced in glycolytic pathway. Not surprisingly, the PTS overexpression bacteria was almost unable to absorb glucose.

Fig 3. The cell growth overnight and glucose uptake of E. coli expressing the PTS transporter in 4 hours
Fig. 4 represented that the cell growth of E. coli expressing the Na+/Glucose transporter was comparable and even slightly higher than the control group. The glucose began to be absorbed at the 3rd hour. The glucose uptake efficiency was achieved upto 97% in Na+/Glu group and greater than in control group with 1.2 times difference.

Fig 4. The cell growth overnight and glucose uptake of E. coli expressing the Na+/Glu transporter at different time point
Discussion
We’ve successfully genetically engineered E. coli expressing high-affinity active PTS transporter system and low-affinity facilitated Na+/glucose cotransporter system by screening in MacConkey and glycerol agar plate with chloramphenicol, respectively. Our data is consistent with the previous study that overexpressing glucose transporter genes do really harm E. coli partly due to disturbing glycolytic pathways.
PTS expressing bacteria grew difficultly in LB broth but Na+/glucose transporter expressing bacteria grew comparably with general E. coli, suggesting that high-affinity active transporter did interact with sugar-related substances in LB culture media and harm the bacterial survival.
In our results, glucose absorption by Na+/glucose transporter expressing bacteria was achieved to 97% after 4 hours with 1.2 times enhanced efficiency compared to the normal E. coli. To further increase the rate of glucose uptake, one may think about the glucose metabolism or conversion to other materials when entering into the cell.

Imagine an undercover agent exposing his id, who is going to commit suicide by taking a poisonous substance. What components in the substance could be and in what situation the agent would trigger the suiciding process.
Back to the view of a microbial cell, normally, a cell has a programmed suicide or defending mechanism to response to unfavorable environments or to outcompete other organisms.
What have iGEMers done and who are the "killers" ?
Lysis gene [BBa_K117000] created by NTU-Singapore in 2008 encodes Lysis protein which could not only lyse bacterial cell membrane but also activate the endonuclease of Colicin E7 (ColE7). The lysis-colicin is one class of bacteriocins which are produced to response to worsening environmental conditions and outcompete other bacteria1
NucA [BBa_K1159105] created by TU-Munich in 2013 from Staphylococcus aureus produces a thermostable exo- and endo-nuclease that is able to degrade genomic DNAs2. NucA also has a role in the cleavage of extracellular DNAs and preventing biofilm formation.
IMPROVE EXISTING PARTS
Part Name [Number]:
- Pcar, synthetic promoter repressed by CRP [BBa_K861171]
- RBS + PhlF repressor + terminator [BBa_K1725041]
- PhlF repressible promoter + strong RBS + GFP [BBa_K1725001]
Triggering the suicide circuit
In our case of sugar hijacking, ideally, the tiny, living “agents” are supposed to kill themselves in the story to make a happy ending. Therefore, we introduced a glucose responsive elements and a repressor circuit to be connected to suicide genes (lysis and NucA).
Promoter Pcar [BBa_K861171] is a glucose responsive promoter created by WHU-China in 2012. Pcar promoter region was de novo designed with overlapping of CRP and RNA polymerase binding site. The initiation of transcription by RNA polymerase may be hindered by the binding of CRP, which occurs at the formation of cAMP-CRP complex in the low concentration of glucose. In other words, when the amount of glucose is high enough, Pcar would be turned on after the leaving of CPR due to the low concentration of cAMP, and vice versa.
PhlF repressor system contains the repressor PhlF [BBa_K1725041] and the PhlF repressible promoter [BBa_K1725001] created by Glasgow in 2015. PhlF could repress GFP fluorescence intensity by 83-fold according to the study of Glasgow’s work.
Glucose responsive repressor device
We’ve innovated this year a novel glucose responsive repressor system (Pcar-wRBS-PhlF-T-Pr-sRBS-GFP/pSB1C3 [BBa_K2230012]) by connecting these two system and extend the function of them.

Please go to DEMONSTRATION section below to check the function of this device in our experimental results.
- - REFERENCE -
- 1. Amount of colicin release in Escherichia coli is regulated by lysis gene expression of the colicin E2 operon. PLoS One. 2015;10(3):e0119124.
- 2. Characterization of a nuclease produced by Staphylococcus aureus. J Biol Chem. 1967;242(5):1016-20.
Gene Cloning
Our final composite parts (device) were listed in the table below. The basic and composite parts composed of the devices were briefly described in the following.

Problem 1.
– BioBricks are unvailable
The part of PI promoter was not in stock. Therefore, we are unable to request from iGEM HQ. We asked and got the plasmid DNA from team NCKU_Tainan. But unfortunately, we can’t re-transform E. coli with materials we got from them.
Trouble-shooting 1.
– Synthesis of promoter sequences on primers
It’s good news that the glucose responsive promoter region contains only 36 bp (PI and Pcar). Therefore, we designed the sequences on the forward primer. The cloning was going smoothly in the following process. And DNA sequences are all confirmed by sequencing.
Problem 2.
– The part of lysis gene was flanked by a wrong BioBrick Prefix
We’ve finished the gene cloning for lysis genes. Unfortunately, according to our sequencing data, we found a mistake that the previous team added the wrong Prefix in front of the part. The part starts with ATG, but unfortunately, the team took the wrong Prefix “for all the other parts”.
Trouble-shooting 2.
– PCR using a primer with correct Prefix
We designed new primers with the correct Prefix “for the coding region parts” along with a strong RBS / a weak RBS, and performed PCR to get the part. This approach successfully solved the previous problem we met.
Demonstration
Experiment

Result

First of all, we’d like to know how glucose responsive promoters (i.e., PI and Pcar) are induced in response to different concentration of glucose (GROUP 1) as shown in in Fig. 1. RFP intensity will be represented as promoter activity and measured at Ex/Em = 584nm/607nm.
Secondly, the glucose responsive repressor system (GROUP 2) will be tested in response to various concentration of glucose. GFP intensity will be measured at Ex/Em = 488nm/518nm as reporter showing the response of repressible promoter in the presence of glucose.
Finally, the glucose responsive suicide circuit (GROUP 2) will be examined in response to decreasing concentration of glucose. OD600 and cell numbers will be calculated to understand the killing efficacy of suicide circuit in the loss of glucose.
The bacteria carrying the indicated vector were cultured in LB media supplemented with 34 μg/ml of chloramphenicol (Cm) at 37°C overnight. The next day, OD600 was measured and adjusted to 2.5 in M9 minimal media with various concentrations of glucose. The bacteria then were incubated for 4 hours at 37°C. RFP, GFP or OD600 were indicators of promoter activities as mentioned. For suicide assay, the culture media were taken out and diluted 106 times following by spreading onto LB Cm agar plate at 37°C overnight. The third day, the numbers of colonies were counted and bacterial viability was calculated.

Result

The result shown in Fig. 2 indicated that PI promoter has significant activity in LB culture media. However, the activity of Pcar promoter is greater than negative control but much smaller than PI and positive control. It’s consistent with the properties of PI and Pcar promoters just mentioned previously in GENE CLONING section and described previously in Part Resgistry [Part: BBa_K861170] by team WHU-China in 2012 who designed the promoters.
In our experiment as presented in Fig. 3, PI and Pcar promoters just responded to various concentrations of glucose with a very slight dose-dependent increase. This phenomenon didn’t correspond to the data provided by team WHU-China in 2012 and team NCKU_Tainan in 2016. Maybe our measurement was not in an optimized condition. Or the reporter of RFP activity was not sensitive enough to respond this difference.



In the assay for repressor system, the data in Fig. 4 gave the similar results as team Glasgow did in 2015, in which the strong activity of the repressible promoter was significantly repressed in the presence of PhlF repressor.
Furthermore, we modified the expression of PhlF under the glucose responsive promoter (Pcar-PhlF-T-Pr-GFP/pSB1C3 [BBa_K2230012]). And the E. coli carrying this plasmid cultured in LB broth overnight was transferred to M9 minimal media with decreasing concentrations of glucose. The result in Fig 5 clearly indicated that the GFP activity driven by the repressible promoter was gradually increased in response to the loss of glucose to 1.88 folds compared to the initial GFP activity at the beginning culture in M9 media, suggesting that the level of expression of PhlF was positively corresponding to the concentration of glucose.


Fig 4. The repressible promoter activity in the absence or presence of PhlF repressor. E. coli carrying the plasmids were cultured in LB broth with Cm at 37°C overnight.
Fig 5. The repressible promoter activity in response to the expression of PhlF repressor under the glucose responsive promoter at different levels of glucose concentration.
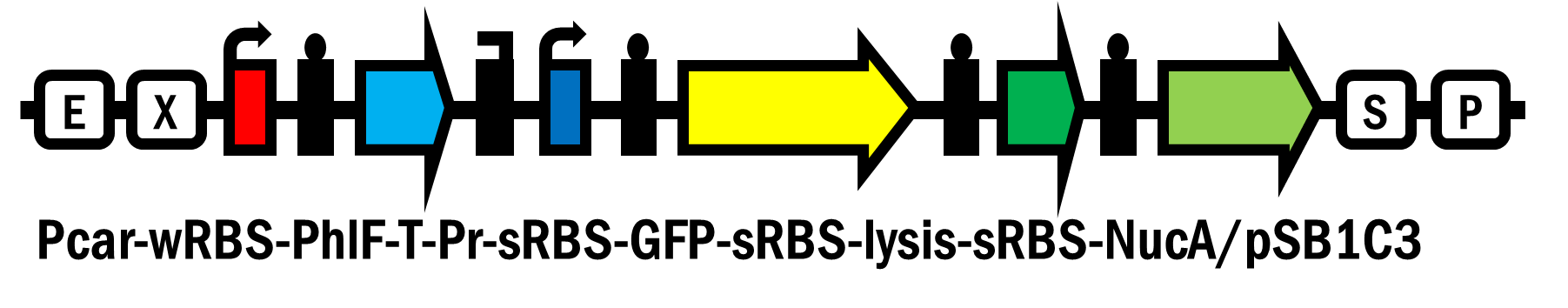
To achieve our goal, we added the suicide genes of lysis and NucA in the back of GFP (Pcar-PhlF-T-Pr-GFP-Lysis-NucA/pSB1C3 [BBa_K2230017]) which are controlled under the same repressible promoter and hope to see the activity of suicide genes could respond against the presence of glucose.
As you can see in Fig 6., the OD value in response to the decreasing concentration of glucose was gradually reduced to 1.89 much less than average 2.71 in control group without suicide gene expression, implying that the suicide proteins killed the cells in the loss of glucose in the environment. Moreover, when the bacteria were grown in M9 media with 0.5mM glucose for 4 hours, the survival rate was decreased to 34% compared to 56% of bacteria without suicide genes (Fig. 7). And the cell numbers were reduced to 671 compared to 1120 of bacteria without suicide genes. Both data confirmed that this suicide device works well and indicated that killing process began when glucose in the media was running out.


Fig 6. The relationship between the glucose and OD600 values of bacteria carrying with (rSuicide) or without (rGFP) suicide genes after cultured in M9 media with various concentration of glucose at 37°C for 4hr.
Fig 7. The cell numbers and bacterial viability were counted and calculated after 106 times diluted culture on LB agar plate at 37°C overnight
Discussion
Glucose responsive promoters can drive RFP but respond slightly in the increasing concentration of glucose in M9 minimal media. However, the device of repressor system works well in response to increasing concentration of glucose which controls the level of expression of PhlF repressors through glucose responsive promoter. The difference responses between the devices may result from the sensitivity of RFP and GFP or the experimental conditions.
The suicide circuit connected the combination of repressor system and glucose responsive promoter were successfully demonstrated in response to the presence of glucose in the environment. We not only confirmed the data of iGEM previous work but also improve the existing parts by extending the function and application of biobricks.

Probiotics in health
To develop food supplements by biological engineering, the host to be engineered needs very carefully to be considered. Probiotics have gained great interest as health benefit. Probiotics not only play a role as immune modulatory in treating allergy or autoimmune disease, but also modulate the gut microbiota in the treatment of metabolic syndrome, obesity and diabetes1.
What iGEMers have done
Probiotics are human friendly and have been used as hosts for many iGEM teams based on the properties of (1) safety, they are natural and exist in your gut and many foods, (2) benefits, they can improve health and prevent disease, and (3) acid resistance, they can pass through gastric acid and reside in your gut.
They have been applied in previous iGEM projects as food supplements (uOttawa: 2009 for cellulose; Uppsala: 2013 for nutritional compounds; UCL: 2016 for antioxidant), as cures for metabolic intolerance (Caltech: 2008 for lactose; UC_Davis: 2009 for gluten; Tuebingen: 2016 for fructose; UIUC Illinois: 2014 for theobromine in puppies), to fight diseases (Stanford: 2009 for autoimmune disorders; Trieste: 2012 for antimicrobial peptide; UIUC_Illinois: 2013 for cardiovascular disease; UT-Dallas: 2014 for infectious diseases; Oxford: 2016 for Wilson’s disease), and to treat or prevent cancer (Arizona_State: 2013 for vaccine delivery system; ATOMS-Turkiye: 2013 for cancer therapy; UPF-CRG_Barcelona: 2016 for colorectal cancer prevention).
Nevertheless, in reality, the major of the projects just worked on E. coli as a proof-of-concept demonstration and still have difficulties to engineer real probiotic strains.
Chassis & Engineering Platform
Escherichia coli Nissle is a nonpathogenic E. coli strain used by some of iGEM teams. The strain was isolated in 1917 by Alfred Nissle and used as probiotics in clinical trials for many gastrointestinal disorders2. However, they don’t have any benefitical properties like natural probiotics like lactic acid bacteria.
Lactobacillus plantarum, Lactobacillus johnsonii and Lactobacillus acidophilus have been mentioned or tried to engineer in few iGEM projects. But there’s no evidence to show how they achieve.
Uppsala successfully engineered Lactobacillus reuteri in 2013. They developed the shuttle vectors based on pSB4C15, in which the replication origin was changed with a broad range replicon from the plasmid pJP059. In addition, they also collected a series of useful biobricks working in probiotics including promoters and antibiotic resistance cassette. The promoter CP29 and erythromycin resistance gene were taken in our project.
TMMU_China is another iGEM team to successfully engineer the probiotic, Lactococcus lactis. Although they demonstrated the method and transformed Lactococcus, they didn’t create any BioBrick parts working on transforming the probiotic.
Our Approach
We have genetically engineered a probiotic which stably expresses the genes of interest. Based on the Uppsala’s method, the probiotic was transformed with shuttle vectors and selected by antibiotics. The bacteria have to maintain the plasmids in the pressure of the existence of antibiotics. Further, the concerns may be addressed with the possibilities of horizontal antibiotic gene transfer.
Lactobacillus acidophilus exists in your gut and is one of the most common probiotics added to your food supplements. Lactobacillus acidophilus strain NCFM has been produced and applied to commercial products and studied in many scientific papers including available full genome sequence information3. In addition, we could obtain them from a local Bioresource Collection and Research Center in Taiwan. Therefore, we decided to choose Lactobacillus acidophilus NCFM as our chassis.
Chromosomal integration are stable for the engineered strain to express heterologous genes. We’d like to design and create the BioBrick vectors for transforming probiotics by inserting genes on chromosome through homologous recombination.
- - REFERENCE -
- 1. Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr Res. 2017;61(1):1318034
- 2. Role and mechanisms of action of Escherichia coli Nissle 1917 in the maintenance of remission in ulcerative colitis patients. World J Gastroenterol. 2016;22(24): 5505–5511.
- 3. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A. 2005;102(11):3906-12
Gene Cloning
To engineer Lactobacillus acidophilus by chromosome integration through homologous recombination, the selection of integration site is very important for successful recombination and not disturbing bacterial internal metabolism.
Based on the method created by Grace L. Douglas and Todd R. Klaenhammer1, the region between slpA gene (LBA0169) stop codon and the terminator was chosen as the intergenic insertion location. The gene of slpA encodes a surface-layer protein with a strong constitutive promoter activity, which can also drive the expression of the inserted gene.

Fig 1. The schematic diagram of gene integration location (modified from Appl Environ Microbiol. 2011)
Recombination Vector - pLBA169
To make pLBA169, the downstream region of LBA0169 and the upstream region of the terminator were amplified by PCR. Then, the two PCR-amplified DNA fragments were ligated to pSB1C3 by tripartite ligation.
Next, CP29-RBS-aeBlue was cut from Part: BBa_K1033280. And this gel isolated DNA fragment was further inserted between two recombination regions on pSB1C3. This process created the standard EcoRI-XbaI-PART-SpeI-PstI assembly position.
To delete the extra SpeI recognition site within the one of the recombination region, we performed site-directed mutagenesis to change one nucleotide of SpeI recognition sequence. The final product has been confirmed by DNA sequencing.
You can refer to the following flow chart of gene cloning of the recombination vector.

Erythromycin Resistance Gene as a Selection Marker

RBS-EmR-TT/pSB1C3
Erythromycin is a common selection marker for engineering lactic acid bacteria2. And teams Uppsala and TMMU_China were using it to select the engineered Lactobacillus with erythromycin resistant strains.
However, the BioBrick parts containing the erythromycin resistance gene are currently unavailable. On the other hand, iGEM HQ is unable to service the request of plasmid carrying erythromycin resistance gene. Maybe iGEM HQ can’t manipulate erythromycin resistant bacteria so far.
Therefore, we decided to clone the erythromycin resistance gene (EmR) by ourselves. We searched the vector carrying EmR and found an iGEM team HKUST in 2010 working with it. We got the pMG36e vector from the team and cloned the EmR gene out onto pSB1C3 followed by assembling RBS (B0034) and double terminator (B0015) parts. These will provide iGEM team this resistance gene as selection marker in the future.
Recombination Vector with Reporter and Selection Marker

RBS-EmR-CP29-RBS-aeBlue/pLBA169
Finally, we assembled the pLBA169 vector with RBS-EmR part as a recombination vector of Lactobacillus acidophilus with the selection marker of erythromycin.
- - REFERENCE -
- 1. Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2011;77(20):7365-71.
- 2. Genetic engineering techniques for lactic acid bacteria: construction of a stable shuttle vector and expression vector for β-glucuronidase. Biotechnol Lett. 2014;36(2):327-35.
Demonstration
Erythromycin
First of all, we have to determine which concentration of erythromycin is suitable for selection of Lactobacillus. We tried to culture Lactobacillus on MRS agar plate with or without 1.25 μg/ml of erythromycin in an anaerobic environment. As the photo showed, Lactobacillus grew on MRS agar plate but can’t grow on the plate with 1.25 μg/ml of erythromycin, which is consistent with the recommended concentration for Lactobacillus selection

Electroporation
To transform Lactobacillus acidophilus with the vector we created, we performed electroporation using BTX Gemini X2 Electroporation System. We followed the protocol provided by BTX but can’t get the recombinant strain yet. At this moment, we’re still try testing in different conditions and hope to achieve success in the future.
Summary
Sugar Crush seems an impossible mission to achieve. Ideally in our mind, the product will absorb glucose in the environment efficiently and compete with small intestine which evolutionarily adapt to take up glucose as much as possible. Secondly, the product will commit suicide after sucking up the glucose and leave the gut without sugar. Finally, the product will be harmless or even benefit human health.
In our project this year, we’ve completed at least and submitted 28 BioBrick basic and composite parts to iGEM community in just several months. We should celebrate by ourselves that we’ve already done the impossible work.
In all of these parts engineered by our hands, our favorite and the most exciting part is that we improved and extending the function of existing parts of glucose responsive promoter (Pcar [BBa_K861171]) and a repressor system (RBS-PhlF-T [BBa_K1725041] and PhlF repressible promoter-RBS-GFP [BBa_K1725001]). It’s useful and a proof-of -concept for our product design to demonstrate the suicide circuit is activated in the loss of glucose in the environment (Pcar-wRBS-PhlF-T-Pr-sRBS-GFP-sRBS-lysis-sRBS-NucA/pSB1C3 [BBa_K2230017]).
And the second is the composite parts of transporter devices (CP29-RBS-aeBlue-RBS-crr-RBS-ptsG-TT/pSB1C3 [BBa_K2230027] and CP29-RBS-aeBlue-RBS-STM1128-TT/pSB1C3 [BBa_K2230028]). We can’t assemble the promoter to drive the genes at first. We didn’t give up and continue trying research the paper and testing with various methods, although we’re advised to change with a regulatory promoter. It’s good news that it eventually comes out we got the recombinant DNA.
The third we love is the Lactobacillus recombination vector (RBS-EmR-CP29-RBS-aeBlue/pLBA169 [BBa_K2230004]). Because we met challenges of unavailable antibiotic resistance gene and an extra site of SpeI, we overcame and tackled the obstacles by acquiring the plasmid carrying the erythromycin resistance gene with the help of iGEM team HKUST’s instructor who have used it in 2010, as well as luckily complete the site-directed mutagenesis in just one experiment.
The fourth we don’t actually love but appreciate our team members’ work are that the parts of glucose responsive promoters (PI [BBa_K861170] and Pcar [BBa_K861171]) ) are neither unavailable from iGEM HQ nor getting plasmids from re-transformed E. coli with the plasmid got from the previous iGEM team. We de novo designed the primer and cloned the parts on pSB1A3 (PI-RBS-RFP-T/pSB1A3 [BBa_K2230005] and Pcar-RBS-RFP-T/pSB1A3 [BBa_K2230006]).
We thank iGEM HQ to provide service of sending BioBrick parts which are not in the Distribution kit (actually we asked 3 times in separate times), and appreciate the favor from team CSMU_NCHU _Taiwan to amplify the genes of transporters of Salmonella for us.
The most challenging and difficult part in our project is performing the functional assays. In testing the glucose responsive promoter in response to various concentrations of glucose, we got in trouble that we can’t repeat the data of previous iGEM works. We tried many conditions and just got slightly increasing response to the increasing concentrations of glucose. Luckily, when analyzing glucose responsive repressor system, we got the corresponding repressible response reversely proportional to the concentrations of glucose. We thought the different responses may result from the sensitivity differences between RFP and GFP. In the suicide circuit assay, it’s a beautiful data demonstrating the suicide circuit activates the killing process when running out the environmental glucose.
In glucose uptake analysis, it is unexpected that the high-affinity active glucose transporter is the worse one for bacterial growth and glucose absorption. Compared to the normal E. coli, the one overexpressing low-affinity facilitated glucose transporter gave a 1.2 times increase and achieved 97% of glucose absorption in 4 hours. The high glucose transport at a time gave a huge stress for the bacteria, in which the toxic metabolite may accumulate due to the overloading of glycolysis pathway. To further speed up the glucose uptake within one hour, we think in the future we can plan a novel route to divert the metabolic pathway to relieve the burden.
In iGEM team uOttawa in 2009, the team worked on another probiotic, Lactobacillus plantarum, to make them convert glucose to cellulose to increase dietary fibre in the gut. Although they seems to just cloned the 4 genes of cellulose synthase from Acetobacter xylium and didn’t get the engineered bacteria, we could extend their idea by combining with our glucose transporter system. Maybe it will make a big breakthrough in the food industry.
In addition, the multifunctional vitamin C is one of the essential and important vitamins that human needs to consume from food. But other mammals could produce vitamin C by themselves because they have whole biochemical synthesis pathway that human being lacks one of them (i.e, L-gulonolactone oxidase). It is intriguing that engineering a probiotic to synthesize vitamin C in the gut will raise interest for future biotechnology. It may be included in our whole picture of engineering probiotics as good food supplements1.
Although we’ve created the standard BioBrick vector for transforming Lactobacillus acidophilus with chromosome integration through homologous recombination. And we also created basic and composite parts of erythromycin resistance gene as an antibiotic selection marker for iGEM community. However, currently we have not successfully engineered Lactobacillus with this vector yet. Based on the paper where the recombination regions were designed, we’re confident and hope that by us or other iGEM teams in one day, transforming probiotics become a standard procedure in addition to E. coli and Bacillus subtilis in synthetic biology field and the iGEM community.
Through our research, we not only learned more about the sugar affecting obesity and diabetes but tackle the problem with synthetic biology and biotechnology. Furthermore, we also increase public awareness and understanding of the relationship between excess glucose uptake and health.
- - REFERENCE -
- 1. The relationship between glucose and vitamin C plays a huge role in health. Natural News. November 18, 2011. By Dr. David Jockers
© iGEM Mingdao 2017. Design: Kevin Li. All rights reserved.


