Zsy5691750 (Talk | contribs) |
Yangteng ou (Talk | contribs) |
||
| Line 523: | Line 523: | ||
<div id="pic_fortyseven" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/1/17/Chenxinyuyuantu6.jpg"><br/>Fig 2-9. Mating successfullycolonies</p></div> | <div id="pic_fortyseven" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/1/17/Chenxinyuyuantu6.jpg"><br/>Fig 2-9. Mating successfullycolonies</p></div> | ||
| − | <p>These are parts of successful result of mating mentioned above. Two figures one by one correspondence.</p> | + | <p>These are parts of a successful result of mating mentioned above. Two figures one by one correspondence.</p> |
<p>To sum up, mating switcher can be presented in kinds of yeast with different forms. This proves that our Mating switcher is fast, flexible and efficient.</p> | <p>To sum up, mating switcher can be presented in kinds of yeast with different forms. This proves that our Mating switcher is fast, flexible and efficient.</p> | ||
| − | <p>Meantime, we cultured the transformed yeast in several 5mL liquid <i>SC-Leu</i> at 30℃ and 220 rpm for 12 hours ( Take three samples at a time). We used one sample for centrifugation to precipitate the bacterial and the remaining two remained unchanged. The difference is the fluorescence value we need, then we calculated the value of average them. The excitation wavelength is set at 540nm and the emission wavelength is set at 635nm. Hereafters, we measured the yeast concentration at OD<sub>600</sub>. At last, we divided the fluorescence value by OD<sub>600</sub> to normalize the value and the result data | + | <p>Meantime, we cultured the transformed yeast in several 5mL liquid <i>SC-Leu</i> at 30℃ and 220 rpm for 12 hours ( Take three samples at a time). We used one sample for centrifugation to precipitate the bacterial and the remaining two remained unchanged. The difference is the fluorescence value we need, then we calculated the value of average them. The excitation wavelength is set at 540nm and the emission wavelength is set at 635nm. Hereafters, we measured the yeast concentration at OD<sub>600</sub>. At last, we divided the fluorescence value by OD<sub>600</sub> to normalize the value and the result data is as follows. |
</p> | </p> | ||
<div class="zxx_zoom_demo" align="center"> | <div class="zxx_zoom_demo" align="center"> | ||
| Line 547: | Line 547: | ||
<p>Our mating switch plays an important role in many respects, such as including heavy metal treatment and cell signal switching. And we created a novel method to prove the effectiveness of the switch in an intuitive and effective way. The terminator of the first part (PVUVC) terminates the expression of the downstream gene, proving the validity of the switcher, and the second part (PVRVC) creates an evident method of color conversion to determine the state of the switcher.</p> | <p>Our mating switch plays an important role in many respects, such as including heavy metal treatment and cell signal switching. And we created a novel method to prove the effectiveness of the switch in an intuitive and effective way. The terminator of the first part (PVUVC) terminates the expression of the downstream gene, proving the validity of the switcher, and the second part (PVRVC) creates an evident method of color conversion to determine the state of the switcher.</p> | ||
| − | <p>Aiming to increase the vika-vox system efficiency, we let vika enzyme saturate expression, but the efficiency was still relatively low. We hypothesized that this phenomenon was caused by degradation of the vika enzyme in the YPD culture medium. We’d better to change the composition or proportion of YPD ingredients to find out the best culture conditions. We are looking forward to more | + | <p>Aiming to increase the vika-vox system efficiency, we let vika enzyme saturate expression, but the efficiency was still relatively low. We hypothesized that this phenomenon was caused by degradation of the vika enzyme in the YPD culture medium. We’d better to change the composition or proportion of YPD ingredients to find out the best culture conditions. We are looking forward to more research in this field so that we can make this system work better and even perfectly.</p> |
| − | <p>We use the <i>RFP</i> as the reporting protein. But there exists a drawback that it’s detected with expensive device. A more intuitive reporting strategy need to be developed, maybe it can be seen by bare eyes like <i>E.coli</i> in the near future.</p> | + | <p>We use the <i>RFP</i> as the reporting protein. But there exists a drawback that it’s detected with an expensive device. A more intuitive reporting strategy need to be developed, maybe it can be seen by bare eyes like <i>E.coli</i> in the near future.</p> |
<div class="reference"> | <div class="reference"> | ||
| Line 582: | Line 582: | ||
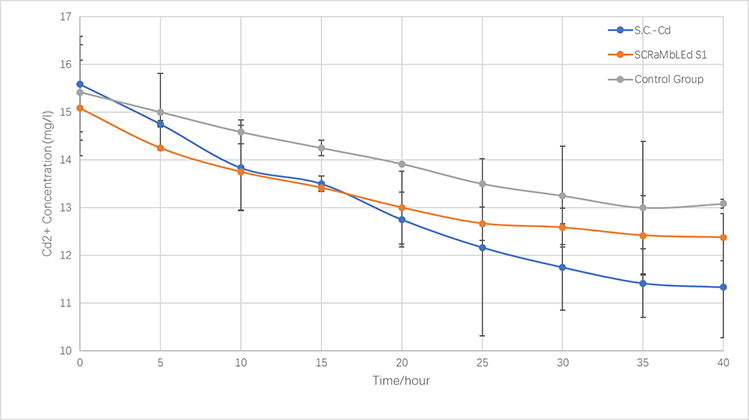
<p>After doing relevant literature reading, we found that yeast’s tolerance level of ambient copper and cadmium ions has a threshold concentration, approximately 3mM and 0.5mM in SC culture media respectively.</p> | <p>After doing relevant literature reading, we found that yeast’s tolerance level of ambient copper and cadmium ions has a threshold concentration, approximately 3mM and 0.5mM in SC culture media respectively.</p> | ||
<p> In order to increase yeast strains’ inherent tolerance of copper or/and cadmium ions in their growing environment, we used this cutting-edge biological technology—SCRaMbLE, which stands for Synthetic Chromosome Rearrangement and Modification by <i>Loxpsym</i>-mediated Evolution, to obtain yeast strain with better tolerance to heavy metal ions .<sup>[1]</sup>. </p> | <p> In order to increase yeast strains’ inherent tolerance of copper or/and cadmium ions in their growing environment, we used this cutting-edge biological technology—SCRaMbLE, which stands for Synthetic Chromosome Rearrangement and Modification by <i>Loxpsym</i>-mediated Evolution, to obtain yeast strain with better tolerance to heavy metal ions .<sup>[1]</sup>. </p> | ||
| − | <p>We constructed three yeast strains namely 079, 160, and 085. They all have a plasmid containing the CRE-EBD sequence and different nutritional labels<sup>[2]</sup>. 079 and 160 strains have URA3 label, 085 strain has HIS label. After proper induction and screening, we successfully obtained mutated 079, 085 and 160 strains that have a manifest growing advantage over control groups when cultured in SC solid media which contain 0.14 mM cadmium ions or 4.8 mM copper ions. We named those mutated strains with increased tolerance capacity of cadmium ions S1, S2, S3, and S4, and as for copper, S5, S6, S7, and S8.</p> | + | <p>We constructed three yeast strains namely 079, 160, and 085. They all have a plasmid containing the CRE-EBD sequence and different nutritional labels<sup>[2]</sup>. 079 and 160 strains have a URA3 label, 085 strain has a HIS label. After proper induction and screening, we successfully obtained mutated 079, 085 and 160 strains that have a manifest growing advantage over control groups when cultured in SC solid media which contain 0.14 mM cadmium ions or 4.8 mM copper ions. We named those mutated strains with increased tolerance capacity of cadmium ions S1, S2, S3, and S4, and as for copper, S5, S6, S7, and S8.</p> |
<p>In order to characterize their increased tolerance of copper or/and cadmium ions, we designed and conducted two different sets of experiments, in both visible and quantitative manner, to test their ability to cope with adverse environmental conditions.</p> | <p>In order to characterize their increased tolerance of copper or/and cadmium ions, we designed and conducted two different sets of experiments, in both visible and quantitative manner, to test their ability to cope with adverse environmental conditions.</p> | ||
<h4>CONSTRUCTION</h4> | <h4>CONSTRUCTION</h4> | ||
| Line 766: | Line 766: | ||
<hr> | <hr> | ||
| − | <p>Based on the <i>CUP1</i> promoter (<a href="http://parts.igem.org/Part:BBa_K2165004">BBa_K2165004</a>) provided by iGEM16_Washington, we constructed this biosensor. To characterize this biosensor, strains of <i>S. cerevisiae BY4742</i> containing the plasmid with an initial OD<sub>600</sub> of 0.1 were grown for 24 hours in SC-URA medium at 30 degrees Celsius, and then were induced with copper sulfate. Samples in different copper concentration were tested with fluorescent spectrophotometer (Hitachi F-2700) after 1, 6, 12, and 24 hours. This protocol was based on the experience used by Waterloo and Washington iGEM teams and amended by our team.</p> | + | <p>Based on the <i>CUP1</i> promoter (<a href="http://parts.igem.org/Part:BBa_K2165004">BBa_K2165004</a>) provided by iGEM16_Washington, we constructed this biosensor. To characterize this biosensor, strains of <i>S. cerevisiae BY4742</i> containing the plasmid with an initial OD<sub>600</sub> of 0.1 were grown for 24 hours in SC-URA medium at 30 degrees Celsius, and then were induced with copper sulfate. Samples in different copper concentration were tested with the fluorescent spectrophotometer (Hitachi F-2700) after 1, 6, 12, and 24 hours. This protocol was based on the experience used by Waterloo and Washington iGEM teams and amended by our team.</p> |
| Line 805: | Line 805: | ||
<hr> | <hr> | ||
| − | <p>The Cu-induced promoter <i>CUP1</i> promoter is a previous BioBrick used by iGEM16_Washington, iGEM16_Waterloo, and other iGEM teams. However, the detailed characterization like what we did this year | + | <p>The Cu-induced promoter <i>CUP1</i> promoter is a previous BioBrick used by iGEM16_Washington, iGEM16_Waterloo, and other iGEM teams. However, the detailed characterization like what we did this year hasn't been shown on iGEM parts page. Moreover, this part hasn’t be improved by any means or in any ways. Under this situation, we plan to work on this promoter to improve its sensitivity and response peak, reduce the leakage expression, and create new parts for future work.</p> |
| Line 840: | Line 840: | ||
</div> | </div> | ||
| − | <p>Based on this mechanism, we redesigned the part sequence provided by iGEM16_Washington. We deleted irrelevant bases on the two ends of this promoter and retained the core sequence. In this way, this promoter played its key role with | + | <p>Based on this mechanism, we redesigned the part sequence provided by iGEM16_Washington. We deleted irrelevant bases on the two ends of this promoter and retained the core sequence. In this way, this promoter played its key role with fewer bases. Strains of <i>S. cerevisiae BY4742</i> containing either BBa_K2165004-yEmRFP and BBa_K2407000-yEmRFP with an initial OD<sub>600</sub> of 0.1 were grown for 24 hours in SC-URA medium at 30 degrees Celsius, and then were induced with 0.1 mM Cu<sup>2+</sup>. Samples were tested with fluorescent spectrophotometer (Hitachi F-2700) after 1, 3, 6, 12, and 24 hours.</p> |
<div class="row"> | <div class="row"> | ||
| Line 874: | Line 874: | ||
<hr> | <hr> | ||
| − | <p>In our experiment, we noticed that <i>CUP1</i> promoter still has a certain degree of leakage expression. To make a better biosensor, we planned to reduce the leakage expression and increase the sensitivity. To reach this goal, we took the fluorescence intensity at both | + | <p>In our experiment, we noticed that <i>CUP1</i> promoter still has a certain degree of leakage expression. To make a better biosensor, we planned to reduce the leakage expression and increase the sensitivity. To reach this goal, we took the fluorescence intensity at both inductions or not into evaluation indexes.</p> |
| − | <p>The technology of error-prone PCR was taken into our experiment. Although there are many methods to introduce genetic diversity into a parent sequence, error-prone PCR is the most common way of creating a combinatorial library based on a single sequence. By adding some heavy metal ions into the PCR buffer and preparing dNTPs with different composition, new mutants were introduced into CUP1 promoter. The whole procedure is shown | + | <p>The technology of error-prone PCR was taken into our experiment. Although there are many methods to introduce genetic diversity into a parent sequence, error-prone PCR is the most common way of creating a combinatorial library based on a single sequence. By adding some heavy metal ions into the PCR buffer and preparing dNTPs with different composition, new mutants were introduced into the CUP1 promoter. The whole procedure is shown in the following figure.</p> |
<div class="row"> | <div class="row"> | ||
| Line 939: | Line 939: | ||
| − | <p>Second, we worked on the sensitivity of biosensor. Leakage expression was not the only thing needed to be solved, and we also needed to increase the response range when it was induced. A good biosensor needs less leakage and more sensitivity. </p> | + | <p>Second, we worked on the sensitivity of the biosensor. Leakage expression was not the only thing needed to be solved, and we also needed to increase the response range when it was induced. A good biosensor needs less leakage and more sensitivity. </p> |
<p>We tested the 4 selected biosensors and control group with 0, 10, 100, 500, and 1000 μM Cu<sup>2+</sup> induced for 20 min, and the result is shown below with logarithmic coordinates.</p> | <p>We tested the 4 selected biosensors and control group with 0, 10, 100, 500, and 1000 μM Cu<sup>2+</sup> induced for 20 min, and the result is shown below with logarithmic coordinates.</p> | ||
| Line 970: | Line 970: | ||
</div> | </div> | ||
| − | <p>The figure shows the response | + | <p>The figure shows the response ranges of biosensors with different promoters within 20 min. For most biosensors, the fluorescence intensity increases as copper ion’s concentration increases from 0 to 100 μM. However, when the concentration exceeds 100 μM, the responses of most biosensor become slow, and the fluorescence intensity decreases. A reasonable explanation is that high concentrations of copper can inhibit the biosensor's response within a short time. </p> |
| − | <p>Fortunately, we still found a biosensor who met the requirements of an excellent biosensor. EP-5 has a less leakage and a higher sensitivity. Its fluorescence intensity is lower than the control group by 17 units with no induction and is higher by 21 units with 100-μM-Cu induction. By aligning the sequence with CUP1 promoter, we found altered bases mainly located at the both sides of UASs and a deletion of one base even occurred between two UASs. We suspected that the change of sensitivity and leakage expression mainly due to the change of | + | <p>Fortunately, we still found a biosensor who met the requirements of an excellent biosensor. EP-5 has a less leakage and a higher sensitivity. Its fluorescence intensity is lower than the control group by 17 units with no induction and is higher by 21 units with 100-μM-Cu induction. By aligning the sequence with the CUP1 promoter, we found altered bases mainly located at the both sides of UASs and a deletion of one base even occurred between two UASs. We suspected that the change of sensitivity and leakage expression mainly due to the change of spatial distribution and the increase of A/T concentration, which both could influence the binding procedure of transcription factors.</p> |
<div class="row"> | <div class="row"> | ||
| Line 1,008: | Line 1,008: | ||
<p>In our characterization of both primary and improved promoters, we found the effect of induction is not as obvious as expected (Previous iGEM team’s results). After reading some references, we found the activation process is related to the acetylation of H3 and H4 located at <i>CUP1</i> promoter, which showed nucleosome reposition and transcription factors binding might be the main reason for the activation. However, our biosensors were ligated on plasmid pRS416, which usually exists in the nucleus in a supercoiled state. There is only little possibility for a plasmid to binds to histones, so the transcription process shows less activation than that on a chromosome.</p> | <p>In our characterization of both primary and improved promoters, we found the effect of induction is not as obvious as expected (Previous iGEM team’s results). After reading some references, we found the activation process is related to the acetylation of H3 and H4 located at <i>CUP1</i> promoter, which showed nucleosome reposition and transcription factors binding might be the main reason for the activation. However, our biosensors were ligated on plasmid pRS416, which usually exists in the nucleus in a supercoiled state. There is only little possibility for a plasmid to binds to histones, so the transcription process shows less activation than that on a chromosome.</p> | ||
| − | <p>In the future, we plan to construct this biosensor on chromosomes to see whether the result will be more positive. Meanwhile, we will continue enlarging the response peak and | + | <p>In the future, we plan to construct this biosensor on chromosomes to see whether the result will be more positive. Meanwhile, we will continue enlarging the response peak and range to improve this biosensor.</p> |
<div class="reference"> | <div class="reference"> | ||
Revision as of 13:47, 30 October 2017
/* OVERRIDE IGEM SETTINGS */