Nagahama
Contents
- 1 Result and Discussion
- 1.1 Identification of yeast of Funazushi
- 1.2 proof of concept
- 1.3 Confirmation whether sod2 were inserted into pYES2 plasmid.
- 1.4 Confirmation that S. cerevisiae expressing sod2 acquired salt tolerance
- 1.5 Confirmation whether carotene synthesis genes (crtYB, crtI, and crtE) and tHMG1 were expressed.
- 1.6 Reference
Result and Discussion
Identification of yeast of Funazushi
Funazushi was touched with a toothpick and streaked in YPD medium to grow yeast colonies. Colony PCR was performed with a primer that amplifies ITS 1 region. We sequenced the PCR products and identified yeasts of Funazushi. From the result of the sequence, the yeast of Funazushi was identified as S. cerevisiae. Therefore we decided to use S. cerevisiae BY 4742 which is commonly used in experiment as yeast to add nutrition in Funazushi.
proof of concept
First, S. cerevisiae BY4742 in order to be dominant species at high salt concentration and low pH,we investigated whether it is necessary to develop a new recombinant S. cerevisiae adapting severe environment. Accoding thesis, it is known that the salt concentration of Funazushi is 4% and pH is 3.7.So we experimented whether S. cerevisiae BY 4742 could grow like S. cerevisiae of Funazushi under this severe environment.
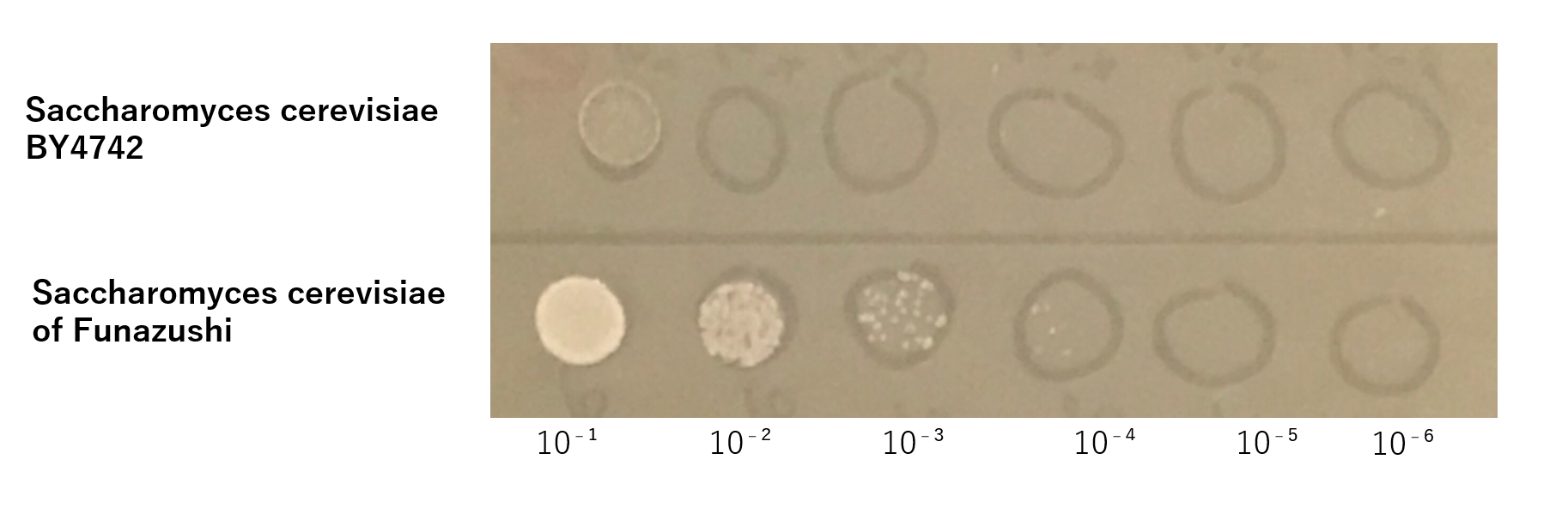
The growth of S. cerevisiae BY4742 cells and S. cerevisiae of Funazushi was tested on plates containing sodium.
This figure showed that S. cerevisiae of Funazushi's growth rate at high salt concentration was faster than S. cerevisiae BY4742.
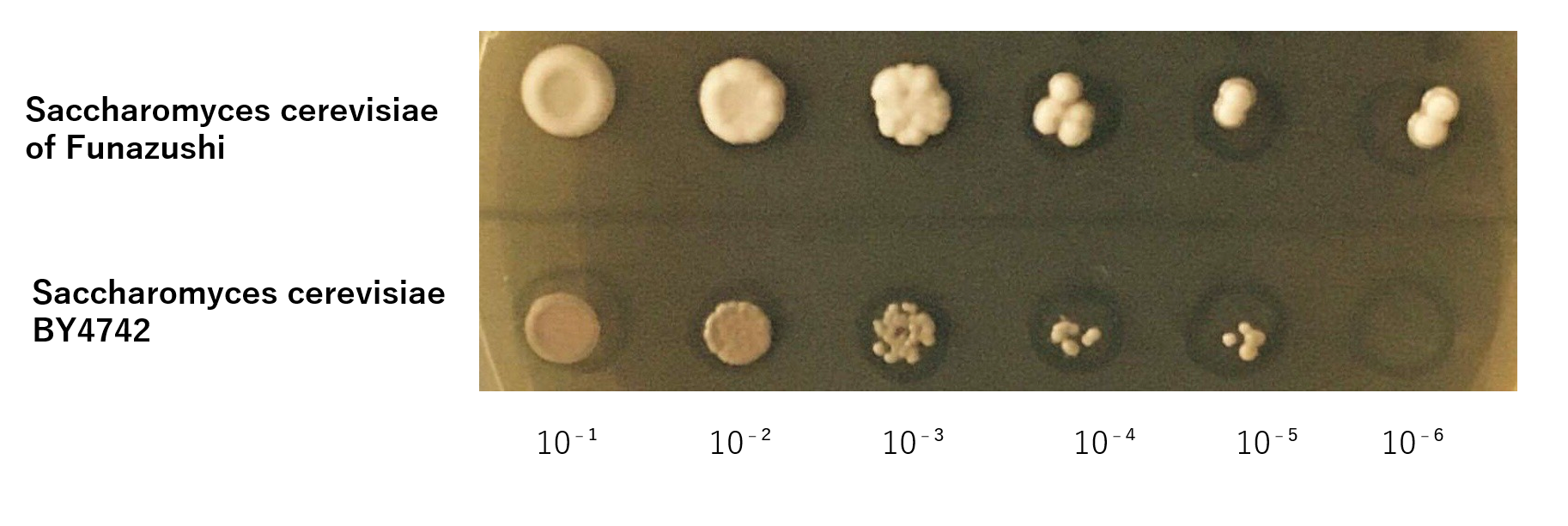
The growth of S. cerevisiae BY4742 cells and S. cerevisiae of Funazushi was tested on plates containing Lactic acid.
This figure showed that S. cerevisiae of Funazushi's growth rate at low pH was faster than S. cerevisiae BY4742.
These results showed that in order to add more nutritional value to Funazushi, we needed to develop new recombinant S. cerevisiae that not only makes more nutrition but also adapts severe environment. So we designed to grow and stand at high salt concentration and low pH.
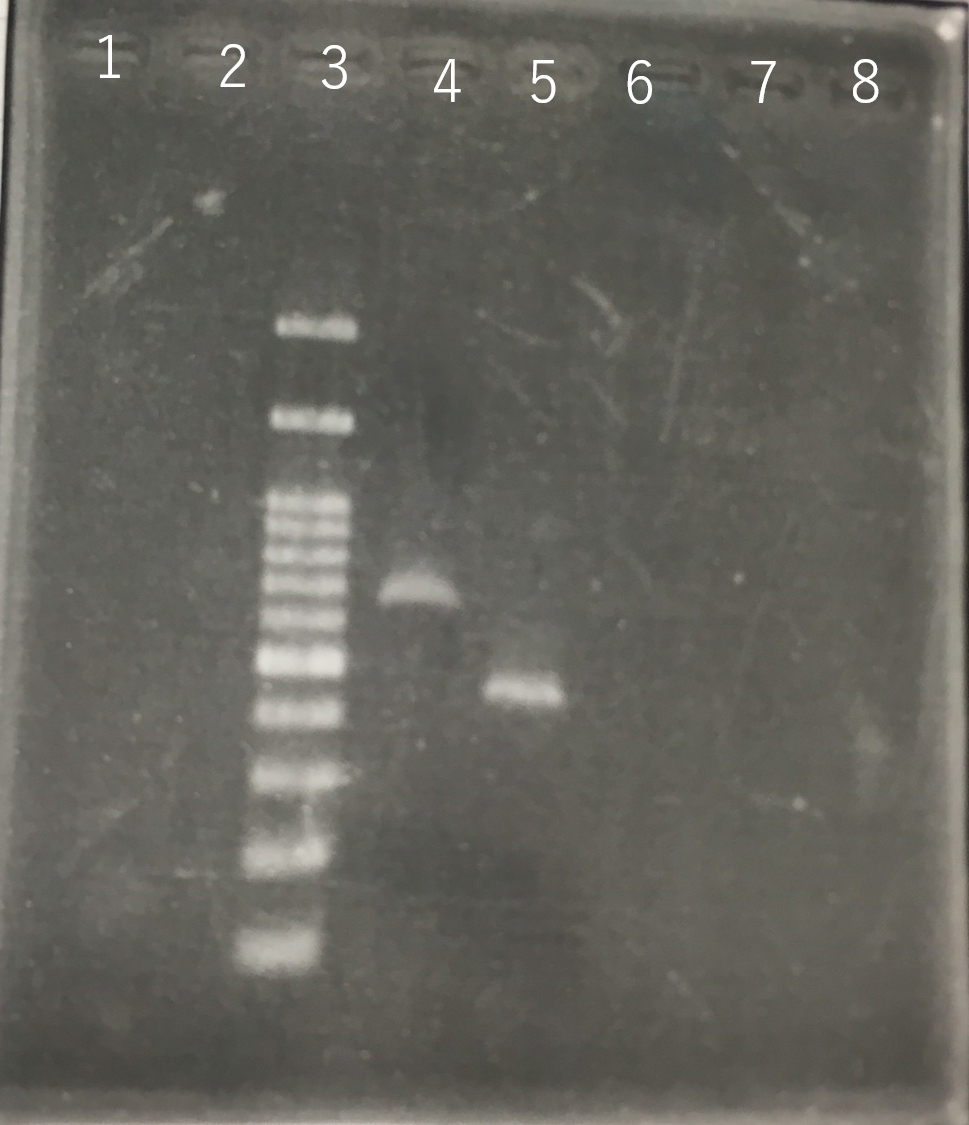
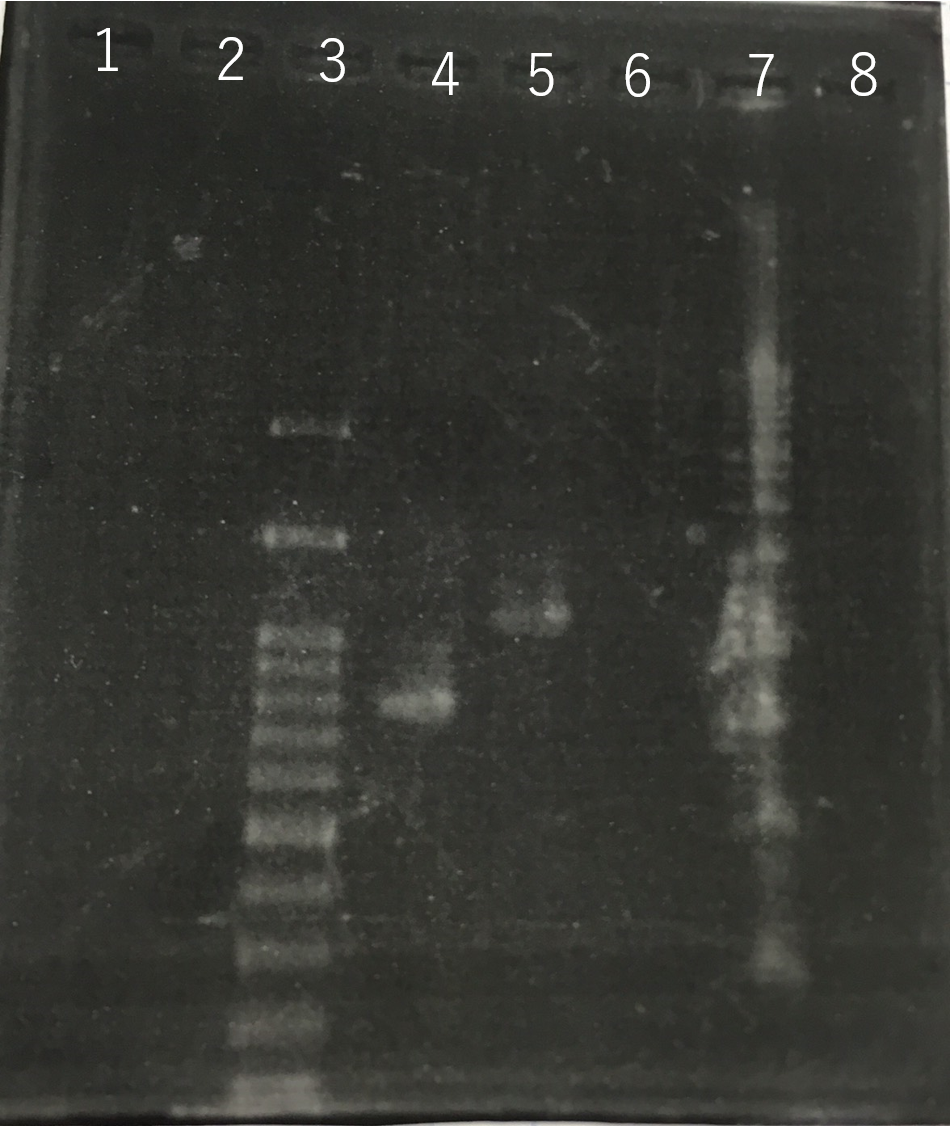
Confirmation whether sod2 were inserted into pYES2 plasmid.
The front and the back part of the intron of sod2 were separately amplified by PCR. The forward primer of sod2 located back the intron contained the sequence of reverse Primer in the front of intron as an adapter.
Fig8.The result of length (the front part of the intron of sod2) by PCR Lane3:100bp DNA ladder Lane4:The front part of the intron of sod2 (185bp)
Using these two PCR products, fusion PCR was performed.
The completed PCR product of the coding region of sod2 was integrated into pYES2 plasmid and transfered the resultant plasmid into E.coli. Plasmid extraction was performed from the transformed E. coli . Using the restriction enzyme used to ligate the PCR product of sod 2 with pYES2 Plasmid, the plasmid extracted from E. coli was cut and the length was confirmed by electrophoresis.
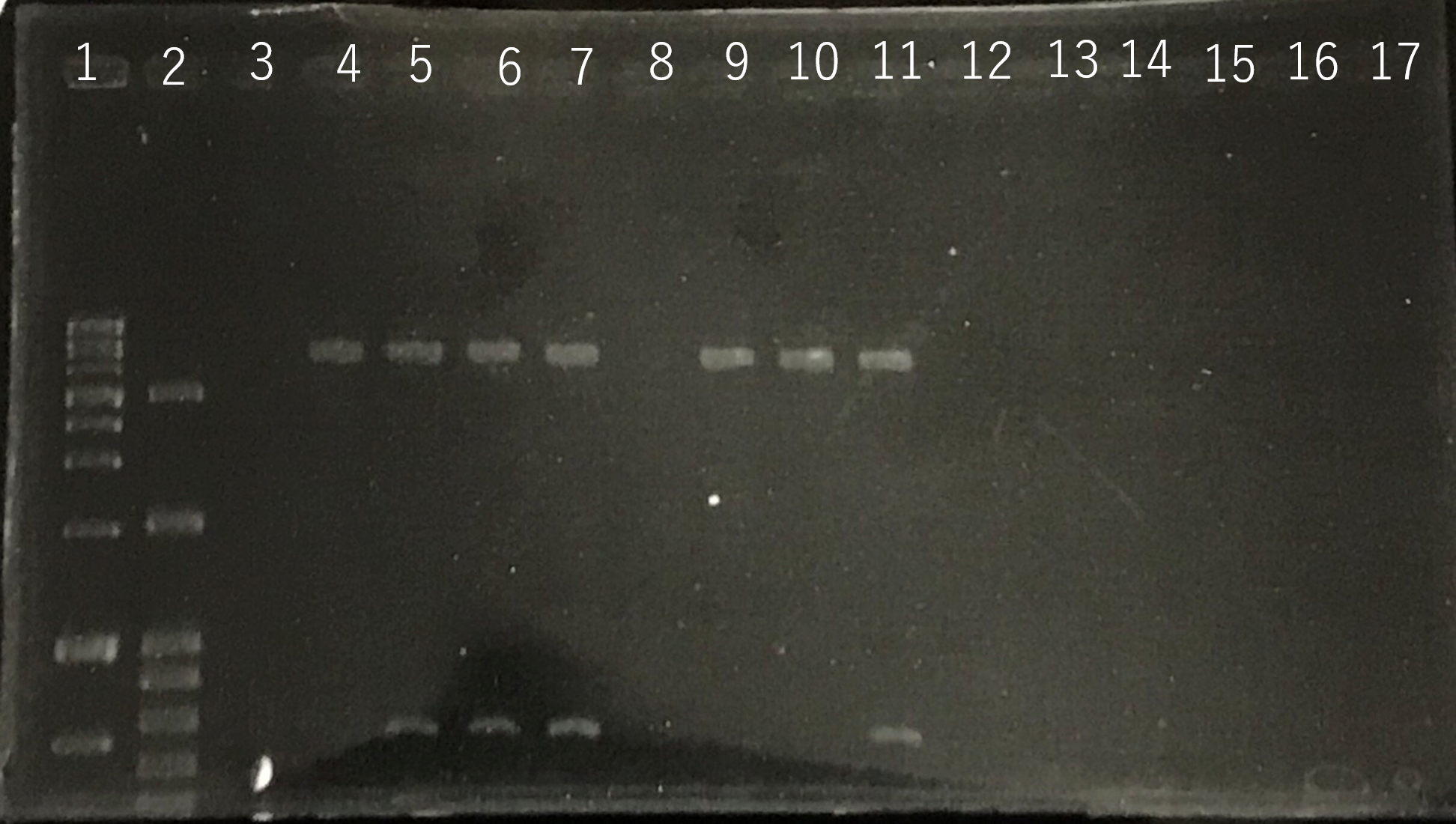
These result showed that sod2 was introduced into pYES2 extracted colony 2, 3, 4 and 8. S. cerevisiae BY4742 was transformed with the plasmid.
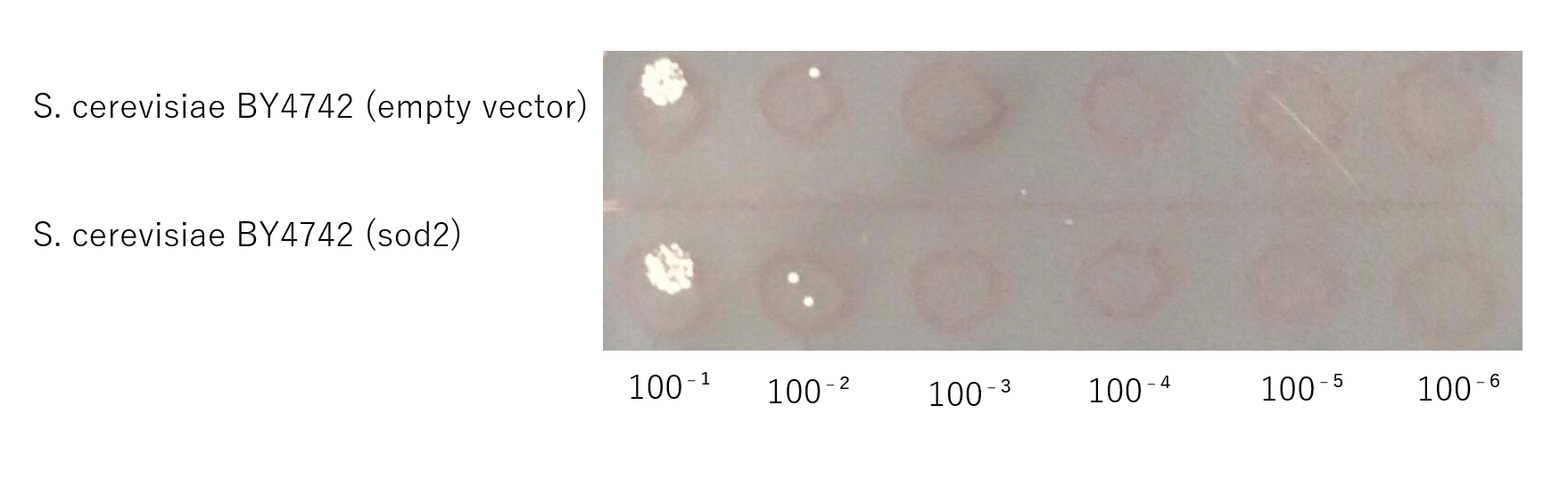
Confirmation that S. cerevisiae expressing sod2 acquired salt tolerance
The growth of S. cerevisiae BY4742 cells expressing sod2 was tested on plates containing sodium.
This figure shows that S. cerevisiae (sod2) cells that overexpress the sod2 product is more survived on SD-ura medium supplemented with 4.0 % (w/v) NaCl than the counterpart control S. cerevisiae transformed with an empty vector.
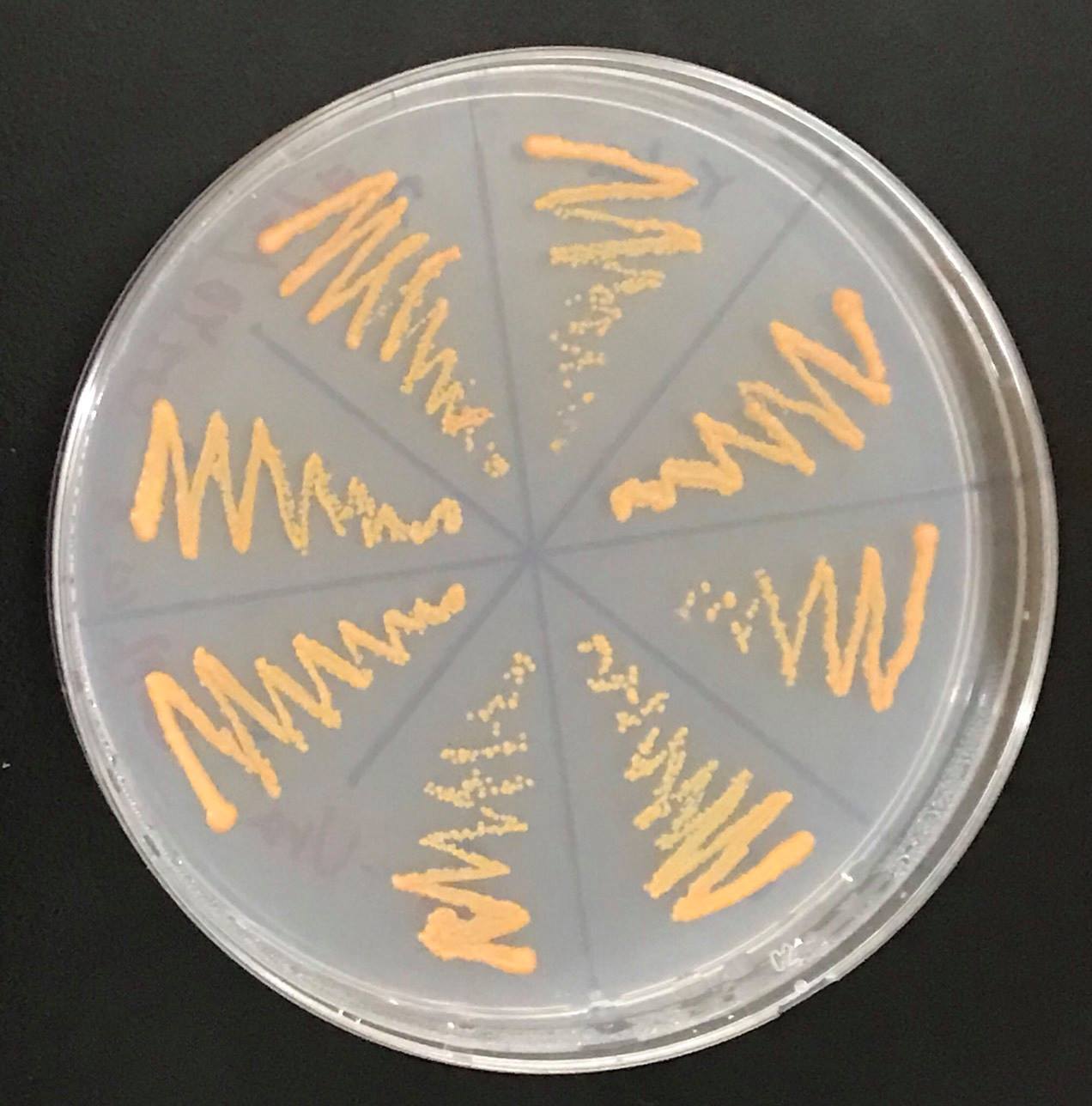
Confirmation whether carotene synthesis genes (crtYB, crtI, and crtE) and tHMG1 were expressed.
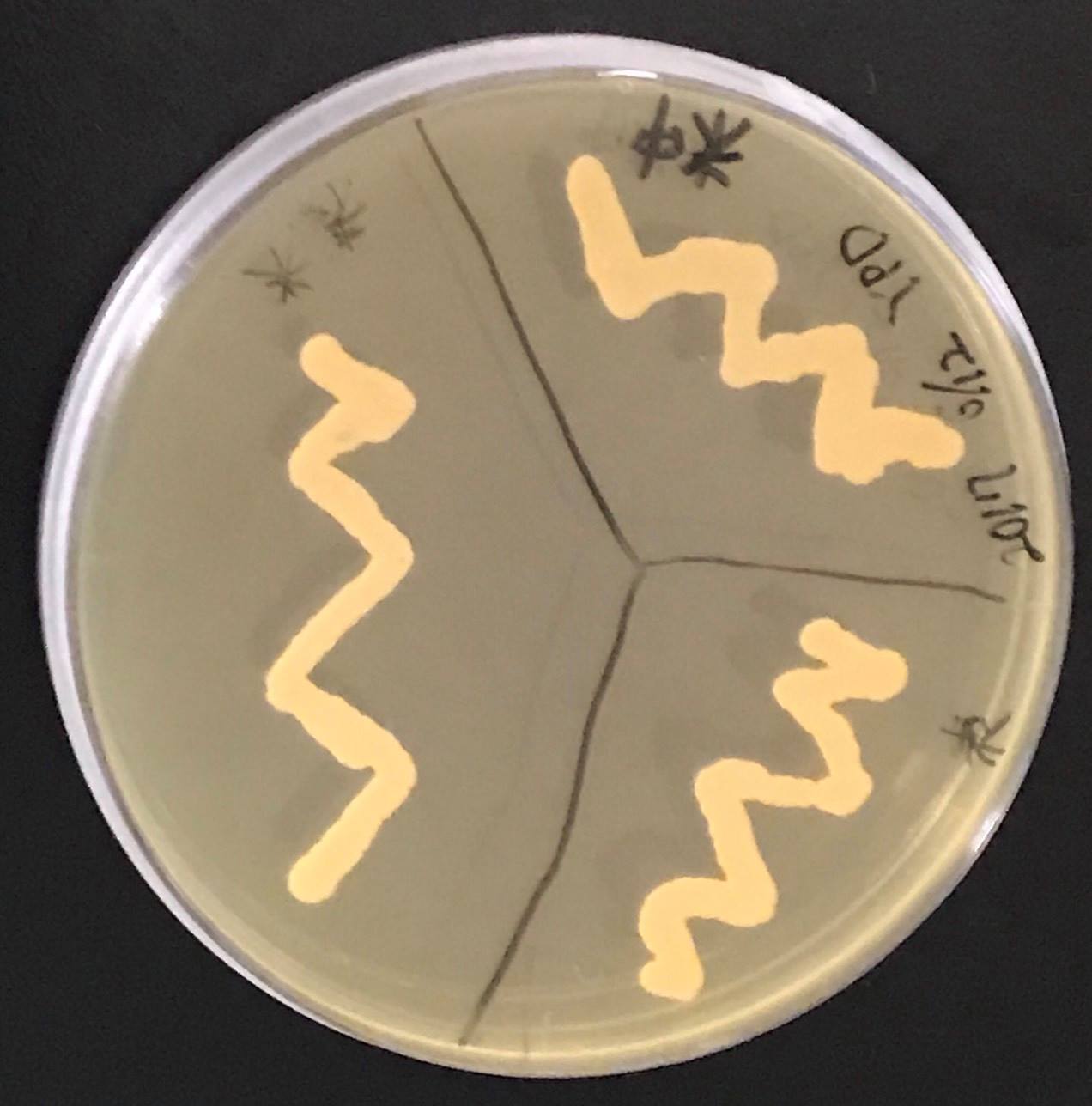
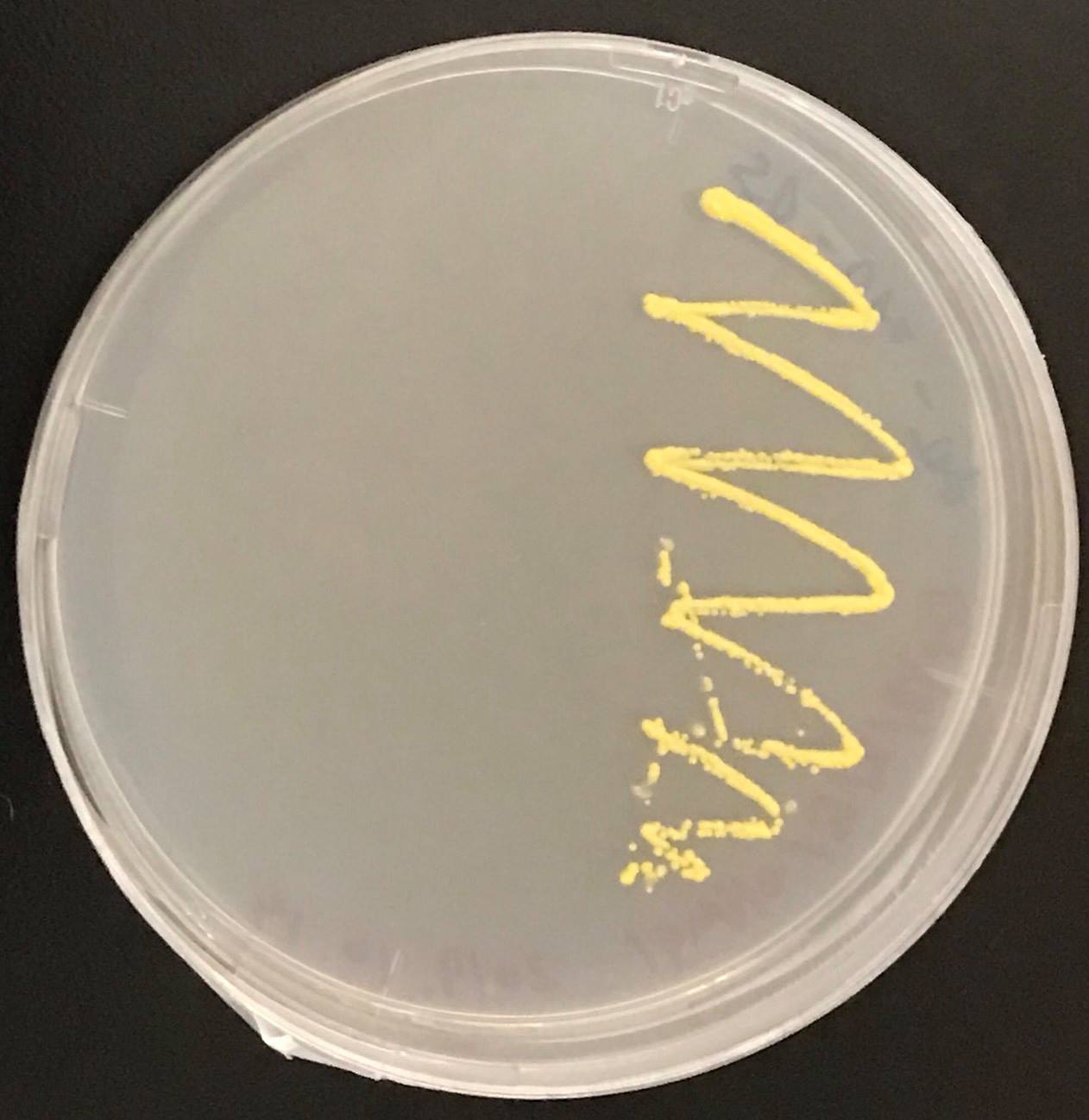
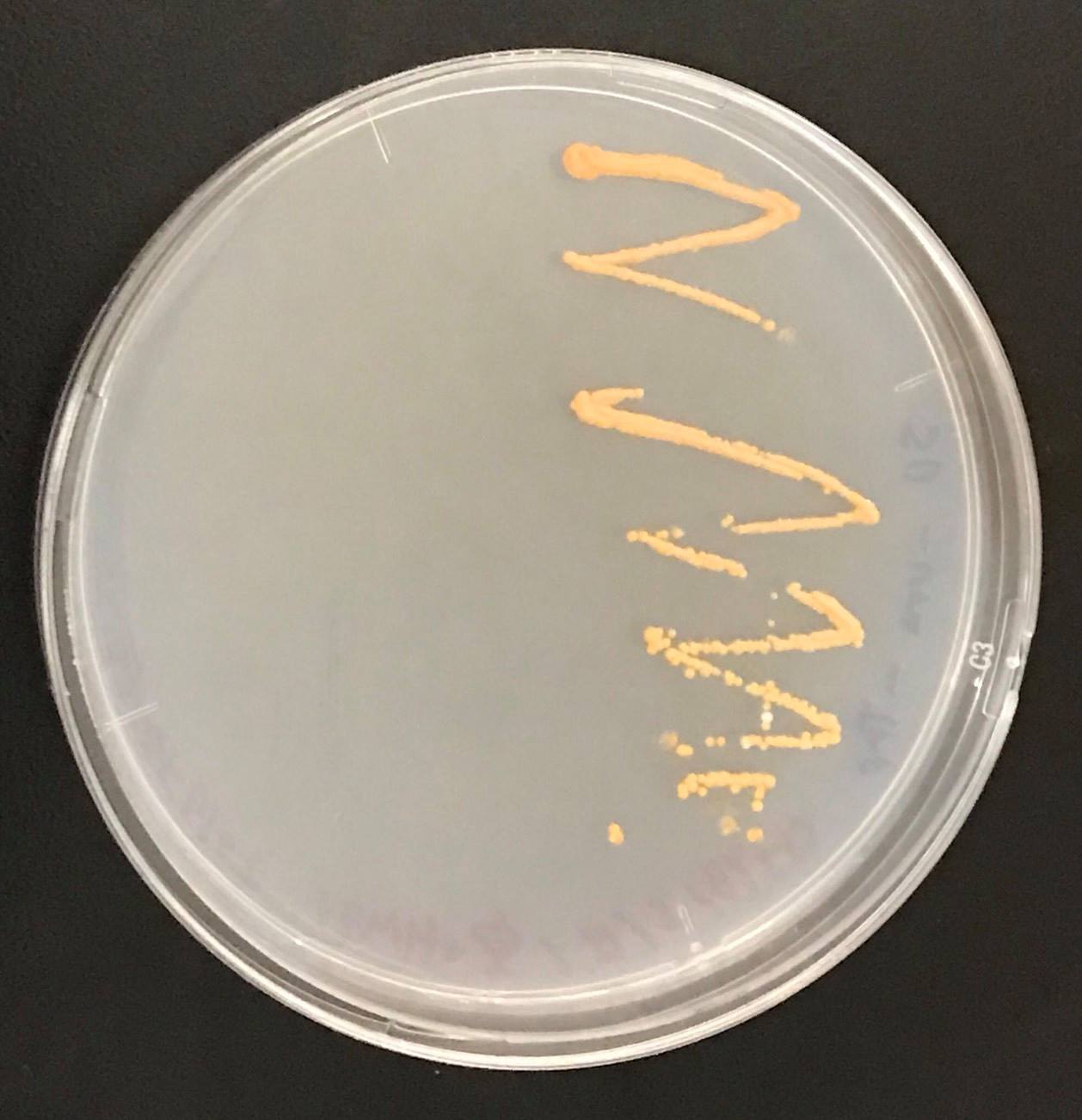
Constructs of carotenogenic genes were inserted on genome of S. cerevisiae. We insertion of crtYB, crtI and crtE resulted in orange colonies.
In addition, in order to increase production of beta-carotene, HMG1 gene was also inserted on the genome of S. cerevisiae. Transformation of tHMG1 resulted in yellow colonies.
These figure shows that color of YB/I/E cells and YB/I/E+tHMG1 cell are clear difference.
We also introduced an empty vector of insert which tHMG1 was not inserted as control S. cerevisiae.
This Color change shows, show that carotene synthesis genes (crtYB, crtI, and crtE) and tHMG1 were expressed.
Accoding to the paper, when tHMG1 is introduced, the amount of β-carotene synthesized is increased seven times as compared with when not. It is thought that S. cerevisiae which we created by inserting YB / I / E + tHMG1 also has increased β carotene content.