| (13 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
<html> | <html> | ||
<head> | <head> | ||
| − | |||
| − | |||
| − | |||
<style> | <style> | ||
| Line 106: | Line 103: | ||
<h1 style="text-align: center; font-size: 4em; font-family: Rubik"> Team Newcastle '17 Presents...</h1> | <h1 style="text-align: center; font-size: 4em; font-family: Rubik"> Team Newcastle '17 Presents...</h1> | ||
<img src="https://static.igem.org/mediawiki/2017/c/c4/T--Newcastle--BB_Logo.png" class="img-fluid rounded mx-auto d-block" style="max-width: 80%" alt=""> | <img src="https://static.igem.org/mediawiki/2017/c/c4/T--Newcastle--BB_Logo.png" class="img-fluid rounded mx-auto d-block" style="max-width: 80%" alt=""> | ||
| − | + | <img src="https://static.igem.org/mediawiki/2017/f/fc/Dec45343.jpeg" class="img-fluid rounded mx-auto d-block" style="max-width: 80%" alt=""> | |
| + | |||
<div class="container-fluid" style="width: 60%"> | <div class="container-fluid" style="width: 60%"> | ||
| − | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">Biosensor genetic networks are typically encoded on a single plasmid within a single chassis. However, this configuration | + | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">Biosensor genetic networks are typically encoded on a single plasmid within a single chassis. However, this configuration means that the biosensor can not be easily modified or re-purposed for new applications.</p> |
<img src="https://static.igem.org/mediawiki/2017/4/42/T--Newcastle--BB_biosensor_plasmid.png" class="img-fluid rounded mx-auto d-block" style="max-width: 40%" alt=""> | <img src="https://static.igem.org/mediawiki/2017/4/42/T--Newcastle--BB_biosensor_plasmid.png" class="img-fluid rounded mx-auto d-block" style="max-width: 40%" alt=""> | ||
| − | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">To alleviate these problems, we propose that biosensor networks are split into three whole-cell modules; a detector, a processor, and a reporter.</p> | + | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">To alleviate these problems, we propose that biosensor networks are split into three whole-cell modules; a detector, a processor, and a reporter. </p> |
<img src="https://static.igem.org/mediawiki/2017/7/75/T--Newcastle--BB_biosensor_modules_abstract.png" class="img-fluid rounded mx-auto d-block" style="max-width: 40%" alt=""> | <img src="https://static.igem.org/mediawiki/2017/7/75/T--Newcastle--BB_biosensor_modules_abstract.png" class="img-fluid rounded mx-auto d-block" style="max-width: 40%" alt=""> | ||
| − | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">With this in mind, we have created the 'Sensynova Biosensor Development Framework'. This framework separates each module into individual cells which communicate via quorum sensing molecules. This separation creates an off-the-shelf set of cellular modules that can be mixed to form new biosensor applications and configurations. This approach also enables biosensor variants to be made and tested without the need for long and tedious genetic cloning steps.</p> | + | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">With this in mind, we have created the 'Sensynova Biosensor Development Framework'. This framework separates each module into individual cells which communicate via quorum sensing molecules. This separation creates an off-the-shelf set of well-characterised cellular modules that can be mixed to form new biosensor applications and configurations. This approach also enables biosensor variants to be made and tested without the need for long and tedious genetic cloning steps. Mixing different ratios of the modules allow the response characteristics of the sensor to be tuned systematically and easily. </p> |
| − | <img src="https://static.igem.org/mediawiki/2017/ | + | <br /> |
| + | <img src="https://static.igem.org/mediawiki/2017/c/ca/T--Newcastle--BB_homepage_image.png" class="img-fluid rounded mx-auto d-block" style="max-width: 100%" alt=""> | ||
| + | <br /> | ||
<p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">We have also included an optional 'adaptor' module. To learn more about our framework, go to our <a href="https://2017.igem.org/Team:Newcastle/Description">project description</a> page!</p> | <p class="text-justify" style="font-size: 1em; margin-top: 2%; margin-bottom: 2%">We have also included an optional 'adaptor' module. To learn more about our framework, go to our <a href="https://2017.igem.org/Team:Newcastle/Description">project description</a> page!</p> | ||
| Line 126: | Line 126: | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
</body> | </body> | ||
</html> | </html> | ||
| + | {{Newcastle_footer}} | ||
Latest revision as of 22:40, 16 November 2017
spacefill
spacefill
Team Newcastle '17 Presents...


Biosensor genetic networks are typically encoded on a single plasmid within a single chassis. However, this configuration means that the biosensor can not be easily modified or re-purposed for new applications.

To alleviate these problems, we propose that biosensor networks are split into three whole-cell modules; a detector, a processor, and a reporter.
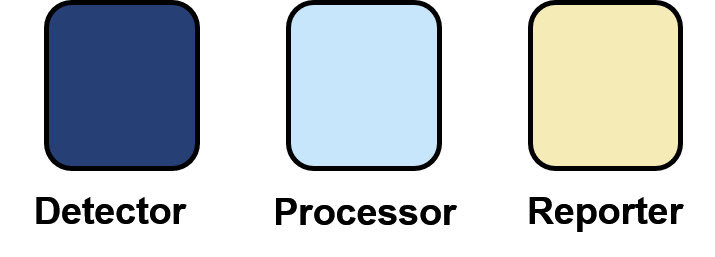
With this in mind, we have created the 'Sensynova Biosensor Development Framework'. This framework separates each module into individual cells which communicate via quorum sensing molecules. This separation creates an off-the-shelf set of well-characterised cellular modules that can be mixed to form new biosensor applications and configurations. This approach also enables biosensor variants to be made and tested without the need for long and tedious genetic cloning steps. Mixing different ratios of the modules allow the response characteristics of the sensor to be tuned systematically and easily.

We have also included an optional 'adaptor' module. To learn more about our framework, go to our project description page!
Welcome to a New Era of

#freethecanary
 |
 |
 |
 |
 |
 |
 |
 |

