| Line 375: | Line 375: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | ||
| − | <p>To ensure the codon usage of our SOX protein was not differing significantly from the average codon usage of <i> E. coli</i>, rare codons were removed from the sequence using the <a href="https://www.idtdna.com/CodonOpt">IDT codon optimisation tool</a> to produce high protein expression. | + | <p>To ensure the codon usage of our SOX protein was not differing significantly from the average codon usage of <i> E. coli</i>, rare codons were removed from the sequence using the<a href="https://www.idtdna.com/CodonOpt">IDT codon optimisation tool</a> to produce high protein expression. |
</br></br> | </br></br> | ||
<i> E. coli</i> BL21-DE3 cells have higher levels of protein expression than DH5α cells and so were a more practical choice. This led to the expression of SOX being placed under the control of a T7 promoter due to BL21-DE3 cells producing T7 polymerase after the addition of IPTG. | <i> E. coli</i> BL21-DE3 cells have higher levels of protein expression than DH5α cells and so were a more practical choice. This led to the expression of SOX being placed under the control of a T7 promoter due to BL21-DE3 cells producing T7 polymerase after the addition of IPTG. | ||
| Line 404: | Line 404: | ||
<p>To prepare SOX for testing, cell cultures were grown following this protocol to step 4. Bradley’s CFPS protocol was then followed (link it). SDS-PAGE gel electrophoresis of the samples was done to check for SOX expression. 1 ml of each culture was lysed with lysozyme and incubated at room temperature before being boiled at 100°C for 10 minutes. 20 µl samples were loaded into each lane. At this point, an error was spotted with the size of SOX on the SDS-PAGE gel (Figure 2).</p> | <p>To prepare SOX for testing, cell cultures were grown following this protocol to step 4. Bradley’s CFPS protocol was then followed (link it). SDS-PAGE gel electrophoresis of the samples was done to check for SOX expression. 1 ml of each culture was lysed with lysozyme and incubated at room temperature before being boiled at 100°C for 10 minutes. 20 µl samples were loaded into each lane. At this point, an error was spotted with the size of SOX on the SDS-PAGE gel (Figure 2).</p> | ||
</br></br> | </br></br> | ||
| − | <p>The band | + | <p>The band was approximately 7 kDa too small. It was then discovered that the sequence synthesised as a gBlock was different to the original sequence found online; parts of the sequence were missing. A new gBlock with the correct sequence was synthesised and the above methods for assembly and preparation for testing were repeated (Figure 3).</p> |
</br></br> | </br></br> | ||
<p>To test for the presence of formaldehyde, and to demonstrate this part works, larger cultures were grown following the aforementioned protocols, and the cells harvested, washed and lysed by sonication. 0 µl, 20 µl, 200 µl and 2 ml of Glyphosate at 10 mg/L concentration was added to the cell lysate and incubated at 37°C. Every 2.5 hours the lysate was tested for the presence of formaldehyde with commercial <a href="http://www.sigmaaldrich.com/catalog/product/sial/37072?lang=en®ion=GB">formaldehyde testing strips</a>.</p> | <p>To test for the presence of formaldehyde, and to demonstrate this part works, larger cultures were grown following the aforementioned protocols, and the cells harvested, washed and lysed by sonication. 0 µl, 20 µl, 200 µl and 2 ml of Glyphosate at 10 mg/L concentration was added to the cell lysate and incubated at 37°C. Every 2.5 hours the lysate was tested for the presence of formaldehyde with commercial <a href="http://www.sigmaaldrich.com/catalog/product/sial/37072?lang=en®ion=GB">formaldehyde testing strips</a>.</p> | ||
| Line 474: | Line 474: | ||
<div id="arsenic" class="collapse"> | <div id="arsenic" class="collapse"> | ||
| − | <h2 style="font-size: 1em"> BioBricks used: <a href="http://parts.igem.org/Part:BBa_J33201">BBa_J33201(Edinburgh )</a>, <a href="http://parts.igem.org/Part:BBa_K2205022">BBa_K2205022 (New)</a> | + | <h2 style="font-size: 1em"> BioBricks used: <a href="http://parts.igem.org/Part:BBa_J33201">BBa_J33201(Edinburgh )</a>, <a href="http://parts.igem.org/Part:BBa_K2205022">BBa_K2205022 (New)</a> </h2> |
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| − | <p>The Sensynova multicellular biosensor platform has been developed to overcome the limitations identified by our team | + | <p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper the success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i> E. coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. |
</br></br> | </br></br> | ||
This section of the project is based on testing the modularity of the system by replacing the IPTG detector part of the Sensynova design with different detecting parts. In particular, an Arsenic sensing part will be used.</p> | This section of the project is based on testing the modularity of the system by replacing the IPTG detector part of the Sensynova design with different detecting parts. In particular, an Arsenic sensing part will be used.</p> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
| − | <p>The part <a href="http://parts.igem.org/Part:BBa_J33201">BBa_J33201</a>was made by the Edinburgh team in 2006.</p> | + | <p>The part <a href="http://parts.igem.org/Part:BBa_J33201">BBa_J33201</a> was made by the Edinburgh team in 2006.</p> |
<img src="https://static.igem.org/mediawiki/2017/b/b1/Vava1aa.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/b/b1/Vava1aa.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
| Line 522: | Line 522: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
| − | <p>Brenner, K., Karing, D., Weiss, R. & Arnold, F. (2007) Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium Proc Natl Acad Sci U S A 104(44): 17300 - 17304 de Mora K, Joshi N, Balint BL, Ward FB, Elfick A, French CE. A pH-based biosensor for detection of arsenic in drinking water. Anal Bioanal Chem. 2011 May; 400(4):1031-9. Epub 2011 Mar 27.</p> | + | <p>Brenner, K., Karing, D., Weiss, R. & Arnold, F. (2007) Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium Proc Natl Acad Sci U S A 104(44): 17300 - 17304 </br> de Mora K, Joshi N, Balint BL, Ward FB, Elfick A, French CE. A pH-based biosensor for detection of arsenic in drinking water. Anal Bioanal Chem. 2011 May; 400(4):1031-9. Epub 2011 Mar 27.</p> |
</div> | </div> | ||
| Line 533: | Line 533: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| − | <p>The Sensynova multicellular biosensor platform has been developed to overcome the limitations identified by our team | + | <p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper the success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Output) by three <i>E.coli </i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. |
</br></br> | </br></br> | ||
| − | This section of the project is based on testing the modularity of the system by implementing the biosensor created by the 2017 Evry Paris-Saclay iGEM team into the Sensynova platform as part of our collaboration requirement.</p> | + | This section of the project is based on testing the modularity of the system by implementing the biosensor created by the 2017 Evry Paris-Saclay iGEM team into the Sensynova platform as part of our <a href="https://2017.igem.org/Team:Newcastle/Collaborations#eps_collab">collaboration</a> requirement.</p> |
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
| Line 611: | Line 611: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
<p> | <p> | ||
| − | Sensynova multicellular biosensor platform has been developed to overcome the limitations that hamper | + | Sensynova multicellular biosensor platform has been developed to overcome the limitations that hamper success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. |
<br /><br /> | <br /><br /> | ||
This part can be used within the platform as a processor unit. Real world applications of biosensors are limited by many factors, one of which is that with most biosensors there is not a readout signal showing if the biosensor is working when not in use, i.e that the cells are still alive and have not lost their biosensor phenotypes. This can make them difficult to use, as well as market, since their viability comes into question as well as leading to false negatives/positives. Biosensors which rely on expression of a reporter signal may also suffer from unobserved activation due to weak or inconstant induction. | This part can be used within the platform as a processor unit. Real world applications of biosensors are limited by many factors, one of which is that with most biosensors there is not a readout signal showing if the biosensor is working when not in use, i.e that the cells are still alive and have not lost their biosensor phenotypes. This can make them difficult to use, as well as market, since their viability comes into question as well as leading to false negatives/positives. Biosensors which rely on expression of a reporter signal may also suffer from unobserved activation due to weak or inconstant induction. | ||
| Line 702: | Line 702: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| − | <p>The Sensynova multicellular biosensor platform has been developed to overcome the limitations identified by our team | + | <p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper the success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. |
</br></br> | </br></br> | ||
This section of the project is based on testing the modularity of the system by inserting two different sensitivity tuner constructs between the processing units of the Sensynova platform; <a href="http://parts.igem.org/Part:BBa_K274371">BBa_K274371</a> and <a href="http://parts.igem.org/Part:BBa_K274381">BBa_K274381</a> . | This section of the project is based on testing the modularity of the system by inserting two different sensitivity tuner constructs between the processing units of the Sensynova platform; <a href="http://parts.igem.org/Part:BBa_K274371">BBa_K274371</a> and <a href="http://parts.igem.org/Part:BBa_K274381">BBa_K274381</a> . | ||
Revision as of 01:55, 30 October 2017
spacefill
spacefill


























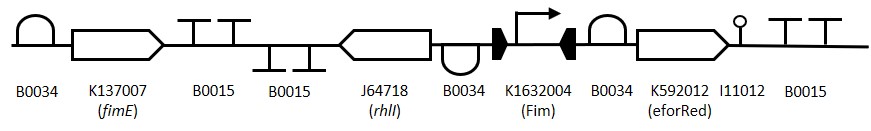
 Figure 2:
Representation of the switching mechanism of the Fim Switch, in the native [OFF] state the eforRED reporter is expressed (shown in red) allowing direct visualisation of the cells. Following the inversion of the promoter region (
Figure 2:
Representation of the switching mechanism of the Fim Switch, in the native [OFF] state the eforRED reporter is expressed (shown in red) allowing direct visualisation of the cells. Following the inversion of the promoter region ( Table 1: Table of parts used for constructing the Fim Switch.
Table 1: Table of parts used for constructing the Fim Switch. Figure 3:
High fidelity amplification of the 3 gBlock fragments for assembly of the Fim Switch. The gBlock-1 amplification is shown in lanes 1+2 (819 bp), gBlock-2 amplification is shown in lanes 3+4 (1148 bp) and the gBlock-3 amplification is shown in lanes 5+6 (939bp).
Figure 3:
High fidelity amplification of the 3 gBlock fragments for assembly of the Fim Switch. The gBlock-1 amplification is shown in lanes 1+2 (819 bp), gBlock-2 amplification is shown in lanes 3+4 (1148 bp) and the gBlock-3 amplification is shown in lanes 5+6 (939bp).
 Figure 4:
Patches of the Fim Switch transformants. Patch number 6 shows the correct red colour which indicates expression of the eforRed chromoprotein.
Figure 4:
Patches of the Fim Switch transformants. Patch number 6 shows the correct red colour which indicates expression of the eforRed chromoprotein.













 The
The