| Line 464: | Line 464: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
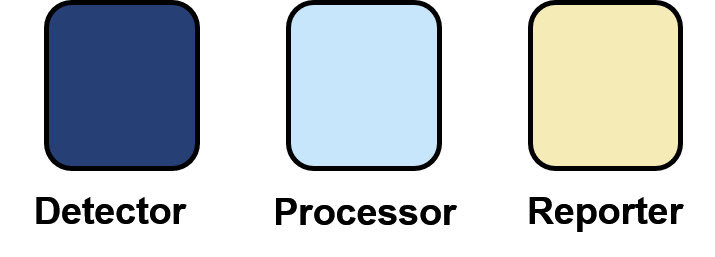
| − | <p>The Sensynova multicellular biosensor platform has been developed to overcome the limitations identified by our team [hyperlink to human practices] that hamper the success in biosensors development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Output) by three E.coli strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. | + | <p>The Sensynova multicellular biosensor platform has been developed to overcome the limitations identified by our team [hyperlink to human practices] that hamper the success in biosensors development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Output) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. |
</br></br> | </br></br> | ||
This section of the project is based on testing the modularity of the system by replacing the IPTG sensing unit present in the Sensynova platform with various synthetic promoters that are regulated by small molecules.</p> | This section of the project is based on testing the modularity of the system by replacing the IPTG sensing unit present in the Sensynova platform with various synthetic promoters that are regulated by small molecules.</p> | ||
| Line 639: | Line 639: | ||
The plasmid backbone was acquired by digestion [Protocol link] of the part K2205015 with XbaI and SpeI, cutting out the original sfGFP construct. | The plasmid backbone was acquired by digestion [Protocol link] of the part K2205015 with XbaI and SpeI, cutting out the original sfGFP construct. | ||
</br></br> | </br></br> | ||
| − | The Psicose detector construct was assembled into the plasmid backbone using the NEB Hi-Fi kit [Protocol link] and transformed into DH5α E. coli cells [Protocol link]. Colony PCR [Protocol link] was performed to check ligations. Colonies picked for this protocol were streaked onto a LB-agar plate. | + | The Psicose detector construct was assembled into the plasmid backbone using the NEB Hi-Fi kit [Protocol link] and transformed into DH5α <i>E. coli</i> cells [Protocol link]. Colony PCR [Protocol link] was performed to check ligations. Colonies picked for this protocol were streaked onto a LB-agar plate. |
</br></br> | </br></br> | ||
Colonies picked from streaked plates and cultures were prepared for miniprepping [Protocol link]. DNA samples were then sent off for sequencing [Website link] to ensure that the constructs were correct.</p> | Colonies picked from streaked plates and cultures were prepared for miniprepping [Protocol link]. DNA samples were then sent off for sequencing [Website link] to ensure that the constructs were correct.</p> | ||
| Line 865: | Line 865: | ||
The part K2205010 contained in pSB1C3, was digested [Protocol link] using SpeI and PstI to allow for the insertion of the processing variants directly after the Las controlled promoter (pLas) that would trigger transcription of sensitivity tuners in the presence of connector 1 of the Sensynova platform. | The part K2205010 contained in pSB1C3, was digested [Protocol link] using SpeI and PstI to allow for the insertion of the processing variants directly after the Las controlled promoter (pLas) that would trigger transcription of sensitivity tuners in the presence of connector 1 of the Sensynova platform. | ||
</br></br> | </br></br> | ||
| − | Ligations were set up overnight [Protocol link] using NEB’s T4 ligase and transformed in DH5α E. coli cells [Protocol link]. Colony PCR [Protocol link] was performed to check ligations. Colonies picked for this protocol were streaked onto a LB-agar plate. | + | Ligations were set up overnight [Protocol link] using NEB’s T4 ligase and transformed in DH5α <i>E. coli</i> cells [Protocol link]. Colony PCR [Protocol link] was performed to check ligations. Colonies picked for this protocol were streaked onto a LB-agar plate. |
</br></br> | </br></br> | ||
Colonies picked from streaked plates and cultures were prepared for miniprepping [Protocol link]. Minipreps were digested [Protocol link] with SpeI and PstI to allow for the insertion of the part K2205011 directly after the PO promoter. | Colonies picked from streaked plates and cultures were prepared for miniprepping [Protocol link]. Minipreps were digested [Protocol link] with SpeI and PstI to allow for the insertion of the part K2205011 directly after the PO promoter. | ||
</br></br> | </br></br> | ||
| − | The part K2205010 contained in pSB1C3, was digested [Protocol link] using XbaI and PstI for BioBrick assembly [Protocol link]. Ligations were set up overnight [Protocol link] using NEB’s T4 ligase and transformed in DH5α E. coli cells [Protocol link]. Colony PCR [Protocol link] was performed to check ligations. Colonies picked for this protocol were streaked onto a LB-agar plate. | + | The part K2205010 contained in pSB1C3, was digested [Protocol link] using XbaI and PstI for BioBrick assembly [Protocol link]. Ligations were set up overnight [Protocol link] using NEB’s T4 ligase and transformed in DH5α <i>E. coli</i> cells [Protocol link]. Colony PCR [Protocol link] was performed to check ligations. Colonies picked for this protocol were streaked onto a LB-agar plate. |
</br></br> | </br></br> | ||
Colonies picked from streaked plates and cultures were prepared for miniprepping [Protocol link]. DNA samples were then sent off for sequencing [Website link] to ensure that the constructs were correct.</p> | Colonies picked from streaked plates and cultures were prepared for miniprepping [Protocol link]. DNA samples were then sent off for sequencing [Website link] to ensure that the constructs were correct.</p> | ||
| Line 916: | Line 916: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | ||
| − | <p>The expression of deGFP was first tested in E. coli cells using an experimental procedure similar to that used in the Interlab study. Cells transformed with pSB1C3-J23100-deGFP were grown in 10 mL LB broth overnight and OD600 nm was measured. Culture was added to 3 separate falcon tubes and made up to 12 mL with LB with chloramphenicol such that the starting OD600 of the culture was approximately 0.02. This set-up was repeated with cells containing an identical plasmid and construct, except sfGFP was in place of deGFP. As a control, untransformed cells were also prepared identically except the LB did not contain chloramphenicol. Tubes with only LB and LB+chloramphenicol were also prepared as blanks. | + | <p>The expression of deGFP was first tested in <i>E. coli</i> cells using an experimental procedure similar to that used in the Interlab study. Cells transformed with pSB1C3-J23100-deGFP were grown in 10 mL LB broth overnight and OD600 nm was measured. Culture was added to 3 separate falcon tubes and made up to 12 mL with LB with chloramphenicol such that the starting OD600 of the culture was approximately 0.02. This set-up was repeated with cells containing an identical plasmid and construct, except sfGFP was in place of deGFP. As a control, untransformed cells were also prepared identically except the LB did not contain chloramphenicol. Tubes with only LB and LB+chloramphenicol were also prepared as blanks. |
</br></br> | </br></br> | ||
The cultures were shake-incubated at 37oC. 300 μL samples from each tube were taken at time points of 15 mins, 2 hours, 4 hours, and 6 hours and stored at 4oC until the end of the experiment. 100 μL of each sample were then added to a 96-well plate. Fluorescence (excitation 485 nm, emission 510 nm) and absorbance (OD600 nm) were measured using a BMG-Labtech fluostar optima plate reader. Fluorescence intensity and grow rates for all three cell types were calculated over time (Figure 1). It was found that while cells expressing sfGFP had a much higher fluorescence intensity than cells expressing deGFP, the growth rate for cells with deGFP was closer to that of untransformed cells. This suggests that in vivo, either deGFP has lower expression than sfGFP, or each molecule of deGFP emits less fluorescence. | The cultures were shake-incubated at 37oC. 300 μL samples from each tube were taken at time points of 15 mins, 2 hours, 4 hours, and 6 hours and stored at 4oC until the end of the experiment. 100 μL of each sample were then added to a 96-well plate. Fluorescence (excitation 485 nm, emission 510 nm) and absorbance (OD600 nm) were measured using a BMG-Labtech fluostar optima plate reader. Fluorescence intensity and grow rates for all three cell types were calculated over time (Figure 1). It was found that while cells expressing sfGFP had a much higher fluorescence intensity than cells expressing deGFP, the growth rate for cells with deGFP was closer to that of untransformed cells. This suggests that in vivo, either deGFP has lower expression than sfGFP, or each molecule of deGFP emits less fluorescence. | ||
| Line 927: | Line 927: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/9/9e/T--Newcastle--BB_in_vivo_deGFP.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/9/9e/T--Newcastle--BB_in_vivo_deGFP.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure 1:</strong> a) fluorescence intensity of E. coli cells overtime. Each bar represents an average of three repeats, and error bars represent +/- standard error. Fluorescence intensity was negative corrected to LB media. b) growth rate of E. coli cells over time. Each bar represents an average of three repeats, and error bars represent +/- standard error. OD¬600 was negative corrected to LB media.</p> | + | <p class="legend"><strong>Figure 1:</strong> a) fluorescence intensity of <i>E. coli</i> cells overtime. Each bar represents an average of three repeats, and error bars represent +/- standard error. Fluorescence intensity was negative corrected to LB media. b) growth rate of <i>E. coli</i> cells over time. Each bar represents an average of three repeats, and error bars represent +/- standard error. OD¬600 was negative corrected to LB media.</p> |
</div> | </div> | ||
| Line 998: | Line 998: | ||
<h4 style="font-family: Rubik; text-align: left; margin-top: 1%">BBa_K1033915 – amajLime</h4> | <h4 style="font-family: Rubik; text-align: left; margin-top: 1%">BBa_K1033915 – amajLime</h4> | ||
| − | <p>The amajLime protein is a yellow-green chromoprotein extracted from the coral Anemonia majano. It was first extracted and characterized by Matz et al. under the name amFP486 (UniProtKB/Swiss-Prot: Q9U6Y6.1 GI: 56749103 GenBank: AF168421.1) and codon optimized for E coli by Genscript. The protein has an absorption maximum at 458 nm giving it a yellow-green colour visible to the naked eye.</p> | + | <p>The amajLime protein is a yellow-green chromoprotein extracted from the coral Anemonia majano. It was first extracted and characterized by Matz et al. under the name amFP486 (UniProtKB/Swiss-Prot: Q9U6Y6.1 GI: 56749103 GenBank: AF168421.1) and codon optimized for <i>E coli</i> by Genscript. The protein has an absorption maximum at 458 nm giving it a yellow-green colour visible to the naked eye.</p> |
<img src="https://static.igem.org/mediawiki/2017/c/ca/T--Newcastle--Lais--amajLime.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/c/ca/T--Newcastle--Lais--amajLime.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
<h4 style="font-family: Rubik; text-align: left; margin-top: 1%">BBa_K1033925 – spisPink</h4> | <h4 style="font-family: Rubik; text-align: left; margin-top: 1%">BBa_K1033925 – spisPink</h4> | ||
| − | <p>The spisPink protein is a pink chromoprotein extracted from the coral Stylophora pistillata. It was first extracted and characterized by Alieva et al. under the name spisCP (GenBank: ABB17971.1) and codon optimized for E. coli by Genscript. The protein has an absorption maximum at 560 nm giving it a pink colour visible to the naked eye. The strong colour is readily observed in both LB or on agar plates after less than 24 hours of incubation.</p> | + | <p>The spisPink protein is a pink chromoprotein extracted from the coral Stylophora pistillata. It was first extracted and characterized by Alieva et al. under the name spisCP (GenBank: ABB17971.1) and codon optimized for <i.E. coli</i> by Genscript. The protein has an absorption maximum at 560 nm giving it a pink colour visible to the naked eye. The strong colour is readily observed in both LB or on agar plates after less than 24 hours of incubation.</p> |
<img src="https://static.igem.org/mediawiki/2017/d/d9/T--Newcastle--Lais--Pink.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/d/d9/T--Newcastle--Lais--Pink.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
<h4 style="font-family: Rubik; text-align: left; margin-top: 1%">BBa_K1033929 – aeBlue</h4> | <h4 style="font-family: Rubik; text-align: left; margin-top: 1%">BBa_K1033929 – aeBlue</h4> | ||
| − | <p>The aeBlue protein is a blue chromoprotein extracted from the basal disk of a beadlet anemone Actinia equine. It was first extracted and characterized by Shkrob et al. 2005 under the name aeCP597 and codon optimised for E. coli by Bioneer Corp. The protein has an absorption maximum at 597nm and a deep blue colour visible to the naked eye. The protein aeBlue has significant sequence homologies with proteins in the GFP family. The coding sequence for this protein was originally submitted to the registry as BBa_K1033916 by the 2012 Uppsala iGEM team.</p> | + | <p>The aeBlue protein is a blue chromoprotein extracted from the basal disk of a beadlet anemone Actinia equine. It was first extracted and characterized by Shkrob et al. 2005 under the name aeCP597 and codon optimised for <i>E. coli</i> by Bioneer Corp. The protein has an absorption maximum at 597nm and a deep blue colour visible to the naked eye. The protein aeBlue has significant sequence homologies with proteins in the GFP family. The coding sequence for this protein was originally submitted to the registry as BBa_K1033916 by the 2012 Uppsala iGEM team.</p> |
<img src="https://static.igem.org/mediawiki/2017/1/1c/T--Newcastle--Lais--Blue.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/1/1c/T--Newcastle--Lais--Blue.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
| Line 1,035: | Line 1,035: | ||
</p></br> | </p></br> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | ||
| − | <p>The chromoproteins aeBlue (BBa_K1033929), amajLime (BBa_K1033915) and spisPink (BBa_K1033925) parts were requested from the iGEM parts registry. Upon arrival, parts were transformed in DH5α E. coli cells [Protocol link]. Colonies were picked and overnight cultures were prepared for miniprepping [Protocol link]. Minipreps were digested [Protocol link] with XbaI and PstI for BioBrick assembly [Protocol link]. | + | <p>The chromoproteins aeBlue (BBa_K1033929), amajLime (BBa_K1033915) and spisPink (BBa_K1033925) parts were requested from the iGEM parts registry. Upon arrival, parts were transformed in DH5α <i>E. coli</i> cells [Protocol link]. Colonies were picked and overnight cultures were prepared for miniprepping [Protocol link]. Minipreps were digested [Protocol link] with XbaI and PstI for BioBrick assembly [Protocol link]. |
</br></br> | </br></br> | ||
The part K2205013 contained in pSB1C3, was digested [Protocol link] using SpeI and PstI to allow for the insertion of the chromoproteins directly after the RhI controlled promoter (pRhI) that would trigger transcription of colour proteins in the presence of connector 2 of the Sensynova platform. | The part K2205013 contained in pSB1C3, was digested [Protocol link] using SpeI and PstI to allow for the insertion of the chromoproteins directly after the RhI controlled promoter (pRhI) that would trigger transcription of colour proteins in the presence of connector 2 of the Sensynova platform. | ||
| Line 1,362: | Line 1,362: | ||
Energy can also be derived from glutamate in the supplement solution (Jewett, et al., 2008), which is added in the form of magnesium glutamate and potassium glutamate. Glutamate is a metabolite in the tricarboxylic acid cycle, which generates NADH. In whole cells, NADH is used in oxidative phosphorylation to produce ATP. Oxidative phosphorylation relies on membrane bound proteins and proton gradients across a membrane. It has been shown previously that extracts prepared using French Press or sonication contain membrane vesicles which have ATPase activity (Futai, 1974), and that oxidative phosphorylation can be activated in CFPS systems (Jewett, et al., 2008). | Energy can also be derived from glutamate in the supplement solution (Jewett, et al., 2008), which is added in the form of magnesium glutamate and potassium glutamate. Glutamate is a metabolite in the tricarboxylic acid cycle, which generates NADH. In whole cells, NADH is used in oxidative phosphorylation to produce ATP. Oxidative phosphorylation relies on membrane bound proteins and proton gradients across a membrane. It has been shown previously that extracts prepared using French Press or sonication contain membrane vesicles which have ATPase activity (Futai, 1974), and that oxidative phosphorylation can be activated in CFPS systems (Jewett, et al., 2008). | ||
</br></br> | </br></br> | ||
| − | Sodium oxalate, another component of the supplement solution, is also used to help increase energy generation by the system. PEP synthetase, an enzyme present in E. coli, converts pyruvate into phosphoenol pyruvate (PEP) in a reaction which consumes ATP, thereby wasting ATP and directing it away from protein synthesis. Oxalate inhibits PEP synthetase by acting as a pyruvate mimic, and hence limit the energy wasted by this reaction. | + | Sodium oxalate, another component of the supplement solution, is also used to help increase energy generation by the system. PEP synthetase, an enzyme present in <i>E. coli</i>, converts pyruvate into phosphoenol pyruvate (PEP) in a reaction which consumes ATP, thereby wasting ATP and directing it away from protein synthesis. Oxalate inhibits PEP synthetase by acting as a pyruvate mimic, and hence limit the energy wasted by this reaction. |
</br></br> | </br></br> | ||
| − | The ribonucleotides ATP, GTP, UTP, and CTP are also components of the supplement solution. They are used in the synthesis of mRNA for transcription of desired genes encoding on exogenous DNA added to the system, and ATP can also be used directly as energy for translation. The polyamines spermidine and putrescine are two other supplements which are added to aid with transcription. It is thought that they can bind proteins and DNA to help recruit polymerase for transcription. Polyamines may also increase translation fidelity, aid ribosome assembly, and activate tRNAs (Jelenc & Kurland, 1979; Jewett & Swartz, 2004b; Algranati & Goldemberg, 1977). To enable translation to occur, amino acids (added separately from the supplement solution) and an E. coli tRNA mixture are added to the CFPS system. Folinic acid is also added as it can be used as a source of folinate for the synthesis of f-Met; the amino acid required for initiation of translation in E. coli. | + | The ribonucleotides ATP, GTP, UTP, and CTP are also components of the supplement solution. They are used in the synthesis of mRNA for transcription of desired genes encoding on exogenous DNA added to the system, and ATP can also be used directly as energy for translation. The polyamines spermidine and putrescine are two other supplements which are added to aid with transcription. It is thought that they can bind proteins and DNA to help recruit polymerase for transcription. Polyamines may also increase translation fidelity, aid ribosome assembly, and activate tRNAs (Jelenc & Kurland, 1979; Jewett & Swartz, 2004b; Algranati & Goldemberg, 1977). To enable translation to occur, amino acids (added separately from the supplement solution) and an <i>E. coli</i> tRNA mixture are added to the CFPS system. Folinic acid is also added as it can be used as a source of folinate for the synthesis of f-Met; the amino acid required for initiation of translation in <i>E. coli</i>. |
</br></br> | </br></br> | ||
Magnesium and potassium ions are also added as supplements. Both ions are ubiquitous in cells with many functions in protein synthesis, namely aiding translation by associating with ribosome subunits and stabilising RNA (Nierhaus, 2014; Pyle, 2002). While magnesium ions are essential for protein synthesis, at high concentrations they can cause inhibition of ribosome translocation and hence inhibit protein synthesis (Borg & Ehrenberg, 2015). </p> | Magnesium and potassium ions are also added as supplements. Both ions are ubiquitous in cells with many functions in protein synthesis, namely aiding translation by associating with ribosome subunits and stabilising RNA (Nierhaus, 2014; Pyle, 2002). While magnesium ions are essential for protein synthesis, at high concentrations they can cause inhibition of ribosome translocation and hence inhibit protein synthesis (Borg & Ehrenberg, 2015). </p> | ||
| Line 1,376: | Line 1,376: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | ||
| − | <p>Cell free extract preparation procedures were based on methods reported in literature previously (Kwon & Jewett, 2015). Cell free extracts were prepared from Escherichia coli BL21 and Bacillus subtilis 168. Cells were streak plated out from glycerol stocks on LB agar (15 mg/mL agar, 10 mg/mL tryptone, 5 mg/mL yeast extract, 0.17 M sodium chloride) and incubated overnight at 37oC. A single colony was used to inoculate 10 mL LB broth (10 mg mL-1 tryptone, 5 mg mL-1 yeast extract, 0.17 M sodium chloride) before shake-incubation at 37oC for approximately 16 hours overnight. 2 mL of overnight liquid culture was used to inoculate 200 mL LB broth in a 2 L flask and shake-incubated at 37oC until late exponential phase was reached (OD600 nm of approximately 2.5 for E. coli BL21 cells). The culture was split in half and cells were harvested by centrifugation at 4,500 RPM and 4oC for 20 minutes in pre-weighed falcon tubes. The wet cell pellet weight was determined before storage at -20oC. Cells were defrosted on ice for approximately 1.5 hours and resuspended in approximately 10 mL of ice-cold CFPS wash buffer (60 mM potassium glutamate, 14 mM magnesium glutamate, 10 mM TRIS (pH 8.2 with acetic acid); autoclave sterilised; supplemented with 2 mM DTT immediately before use) per gram of wet cell pellet. Resuspended cells were centrifuged at 4,500 RPM and 4oC for 20 mins. The supernatant was discarded and cell pellets were resuspended and centrifuged in CFPS wash buffer twice more. The washed pellets were then resuspended in 1 mL CFPS wash buffer per gram of wet cell pellet and aliquoted to 1 mL in 2 mL tubes. Cells were lysed by sonication (20% amplitude, cycles of 40 seconds on – 59.9 seconds off, 432.5 Joules) and the lysates were clarified by centrifugation at 12,000 RPM for 10 mins, flash frozen in liquid nitrogen, and stored at -80oC. A CFPS supplement solution was prepared based on previously reported protocols (Yang, et al., 2012). Amino acid stock solutions were prepared according to Table 1. Briefly, amino acids were weighed in 2 mL tubes, dissolved in 5 M potassium hydroxide, and stored at -20oC. A 10x amino acid solution was prepared by mixing the stock solutions together in amounts according to Table 1, and the pH was adjusted to 7.9 with acetic acid. The solution was aliquoted to 1.5 mL and stored at -80oC. | + | <p>Cell free extract preparation procedures were based on methods reported in literature previously (Kwon & Jewett, 2015). Cell free extracts were prepared from Escherichia coli BL21 and Bacillus subtilis 168. Cells were streak plated out from glycerol stocks on LB agar (15 mg/mL agar, 10 mg/mL tryptone, 5 mg/mL yeast extract, 0.17 M sodium chloride) and incubated overnight at 37oC. A single colony was used to inoculate 10 mL LB broth (10 mg mL-1 tryptone, 5 mg mL-1 yeast extract, 0.17 M sodium chloride) before shake-incubation at 37oC for approximately 16 hours overnight. 2 mL of overnight liquid culture was used to inoculate 200 mL LB broth in a 2 L flask and shake-incubated at 37oC until late exponential phase was reached (OD600 nm of approximately 2.5 for <i>E. coli</i> BL21 cells). The culture was split in half and cells were harvested by centrifugation at 4,500 RPM and 4oC for 20 minutes in pre-weighed falcon tubes. The wet cell pellet weight was determined before storage at -20oC. Cells were defrosted on ice for approximately 1.5 hours and resuspended in approximately 10 mL of ice-cold CFPS wash buffer (60 mM potassium glutamate, 14 mM magnesium glutamate, 10 mM TRIS (pH 8.2 with acetic acid); autoclave sterilised; supplemented with 2 mM DTT immediately before use) per gram of wet cell pellet. Resuspended cells were centrifuged at 4,500 RPM and 4oC for 20 mins. The supernatant was discarded and cell pellets were resuspended and centrifuged in CFPS wash buffer twice more. The washed pellets were then resuspended in 1 mL CFPS wash buffer per gram of wet cell pellet and aliquoted to 1 mL in 2 mL tubes. Cells were lysed by sonication (20% amplitude, cycles of 40 seconds on – 59.9 seconds off, 432.5 Joules) and the lysates were clarified by centrifugation at 12,000 RPM for 10 mins, flash frozen in liquid nitrogen, and stored at -80oC. A CFPS supplement solution was prepared based on previously reported protocols (Yang, et al., 2012). Amino acid stock solutions were prepared according to Table 1. Briefly, amino acids were weighed in 2 mL tubes, dissolved in 5 M potassium hydroxide, and stored at -20oC. A 10x amino acid solution was prepared by mixing the stock solutions together in amounts according to Table 1, and the pH was adjusted to 7.9 with acetic acid. The solution was aliquoted to 1.5 mL and stored at -80oC. |
<br /> | <br /> | ||
</p> | </p> | ||
| Line 1,388: | Line 1,388: | ||
<br /> | <br /> | ||
<p> | <p> | ||
| − | The following solutions were prepared in autoclave sterilised MiliQ water and stored at -80oC: 100x magnesium glutamate solution (1.2 M magnesium glutamate), 10x salt solution (1.3 M potassium glutamate, 40 mM sodium oxalate, 10 mM ammonium acetate), 25x NTPS & co-factor mix (37.5 mM spermidine, 30 mM ATP, 21.25 mM GTP, UTP, and CTP, 25 mM putrescine, 8.25 mM nicotinamide diphosphate, 4.25 mg mL-1 E. coli tRNA (Roche), 0.85 mg mL-1 folinic acid, N xX co-enzyme A), 25x sodium pyruvate solution (825 mM sodium pyruvate, pH to 7.3 with potassium hydroxide), unless stated otherwise. A 5x CFPS supplement solution premix (5% v/v nuclease free water, 5% v/v magnesium glutamate solution, 50% v/v salt solution, 20% v/v NTPS & co-factor mix, 20% v/v sodium pyruvate solution, unless stated otherwise) was prepared and stored at -80oC. CFPS activity of systems prepared as above were tested by expression of 1.7 μg pSB1C3-J23100-sfGFP (Figure 1). Firstly, enough CFPS master mix was prepared for 7 reactions by mixing 112 μL cell extract, 70 μL CFPS supplement premix, and 21 μL amino acid solution in a 1.5 mL tube and stored on ice. A further six 1.5 mL tubes were put on ice; 21 μL of nuclease free water was added to three tubes, and 1.7 μg pSB1C3-J23100-sfGFP plasmid DNA from the same stock solution was added to the remaining three. Tubes containing DNA were made up to 21 μL with nuclease-free water. CFPS master mix (29 μL) was then added to all tubes, which were vortexed and transferred to a 96-well plate. The plate was incubated in a BMG Labtech Fluostar Optima at 370C for 4.25 hours with fluorescence readings (excitation: 485 nm, emission: 510 nm) every 15 mins. Figure 2 shows that over time, fluorescence intensity increased in systems with DNA encoding for sfGFP compared to systems with no DNA. Hence, the system had CFPS activity.</p> | + | The following solutions were prepared in autoclave sterilised MiliQ water and stored at -80oC: 100x magnesium glutamate solution (1.2 M magnesium glutamate), 10x salt solution (1.3 M potassium glutamate, 40 mM sodium oxalate, 10 mM ammonium acetate), 25x NTPS & co-factor mix (37.5 mM spermidine, 30 mM ATP, 21.25 mM GTP, UTP, and CTP, 25 mM putrescine, 8.25 mM nicotinamide diphosphate, 4.25 mg mL-1 <i>E. coli</i> tRNA (Roche), 0.85 mg mL-1 folinic acid, N xX co-enzyme A), 25x sodium pyruvate solution (825 mM sodium pyruvate, pH to 7.3 with potassium hydroxide), unless stated otherwise. A 5x CFPS supplement solution premix (5% v/v nuclease free water, 5% v/v magnesium glutamate solution, 50% v/v salt solution, 20% v/v NTPS & co-factor mix, 20% v/v sodium pyruvate solution, unless stated otherwise) was prepared and stored at -80oC. CFPS activity of systems prepared as above were tested by expression of 1.7 μg pSB1C3-J23100-sfGFP (Figure 1). Firstly, enough CFPS master mix was prepared for 7 reactions by mixing 112 μL cell extract, 70 μL CFPS supplement premix, and 21 μL amino acid solution in a 1.5 mL tube and stored on ice. A further six 1.5 mL tubes were put on ice; 21 μL of nuclease free water was added to three tubes, and 1.7 μg pSB1C3-J23100-sfGFP plasmid DNA from the same stock solution was added to the remaining three. Tubes containing DNA were made up to 21 μL with nuclease-free water. CFPS master mix (29 μL) was then added to all tubes, which were vortexed and transferred to a 96-well plate. The plate was incubated in a BMG Labtech Fluostar Optima at 370C for 4.25 hours with fluorescence readings (excitation: 485 nm, emission: 510 nm) every 15 mins. Figure 2 shows that over time, fluorescence intensity increased in systems with DNA encoding for sfGFP compared to systems with no DNA. Hence, the system had CFPS activity.</p> |
| Line 1,406: | Line 1,406: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/e/e3/T--Newcastle--BB_CFPS_initial_test.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/e/e3/T--Newcastle--BB_CFPS_initial_test.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure 2:</strong> Negative corrected fluorescence for E. coli BL21 extract-based CFPS systems. Each data point is an average of 3 replicate reactions, and error bars represent +/- standard error.</p> | + | <p class="legend"><strong>Figure 2:</strong> Negative corrected fluorescence for <i>E. coli</i> BL21 extract-based CFPS systems. Each data point is an average of 3 replicate reactions, and error bars represent +/- standard error.</p> |
</div> | </div> | ||
| Line 1,433: | Line 1,433: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Experimental Procedure 1</h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Experimental Procedure 1</h2> | ||
| − | <p>Cell extracts were prepared from E. coli BL21 cells using sonication. A CFPS supplement premix solution was prepared as above, except the salts were omitted. Separate solutions for each salt were prepared and added to each CFPS reaction according to the main effects screening design. Reactions were performed as above and CFPS activity was measured as fluorescence at each time point minus fluorescence at 15 mins (Figure 3). Endpoint data was then used, along with the JMP software, to build a model predicting the important factors (Figure 4). | + | <p>Cell extracts were prepared from <i>E. coli</i> BL21 cells using sonication. A CFPS supplement premix solution was prepared as above, except the salts were omitted. Separate solutions for each salt were prepared and added to each CFPS reaction according to the main effects screening design. Reactions were performed as above and CFPS activity was measured as fluorescence at each time point minus fluorescence at 15 mins (Figure 3). Endpoint data was then used, along with the JMP software, to build a model predicting the important factors (Figure 4). |
</br></br> | </br></br> | ||
Revision as of 17:38, 31 October 2017
spacefill
spacefill
Our Experimental Results
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project
































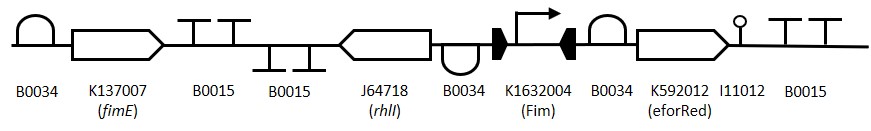
 Figure 2:
Representation of the switching mechanism of the Fim Switch, in the native [OFF] state the eforRED reporter is expressed (shown in red) allowing direct visualisation of the cells. Following the inversion of the promoter region (
Figure 2:
Representation of the switching mechanism of the Fim Switch, in the native [OFF] state the eforRED reporter is expressed (shown in red) allowing direct visualisation of the cells. Following the inversion of the promoter region ( Table 1: Table of parts used for constructing the Fim Switch.
Table 1: Table of parts used for constructing the Fim Switch. Figure 3:
High fidelity amplification of the 3 gBlock fragments for assembly of the Fim Switch. The gBlock-1 amplification is shown in lanes 1+2 (819 bp), gBlock-2 amplification is shown in lanes 3+4 (1148 bp) and the gBlock-3 amplification is shown in lanes 5+6 (939bp).
Figure 3:
High fidelity amplification of the 3 gBlock fragments for assembly of the Fim Switch. The gBlock-1 amplification is shown in lanes 1+2 (819 bp), gBlock-2 amplification is shown in lanes 3+4 (1148 bp) and the gBlock-3 amplification is shown in lanes 5+6 (939bp).
 Figure 4:
Patches of the Fim Switch transformants. Patch number 6 shows the correct red colour which indicates expression of the eforRed chromoprotein.
Figure 4:
Patches of the Fim Switch transformants. Patch number 6 shows the correct red colour which indicates expression of the eforRed chromoprotein.













 The
The