| Line 1,373: | Line 1,373: | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| Line 1,392: | Line 1,389: | ||
<div id="cfps" class="collapse" style="border: 1px solid #222222; padding: 1%"> | <div id="cfps" class="collapse" style="border: 1px solid #222222; padding: 1%"> | ||
| − | <p>Cells extracts being used in CFPS systems tend to be supplemented with a cocktail of compounds and molecules to aid the process of transcription and translation. Although exact supplement solutions can vary from protocol to protocol, most have the same basic composition; salts, nucleotides, tRNAs, co-factors, energy sources, and amino acids (Yang, <i>et al</i>., 2012). The supplement solution used in this study is based on the Cytomin system | + | <p>Cells extracts being used in CFPS systems tend to be supplemented with a cocktail of compounds and molecules to aid the process of transcription and translation (Figure 1). Although exact supplement solutions can vary from protocol to protocol, most have the same basic composition; salts, nucleotides, tRNAs, co-factors, energy sources, and amino acids (Yang, <i>et al</i>., 2012). The supplement solution used in this study is based on the Cytomin system (Jewett, <i>et al</i>., 2008). For the cytomin supplement solution, the major energy source is sodium pyruvate, which is converted to acetate through a series of reactions catalysed by enzymes in the crude cell extract. The first reaction, pyruvate to acetyl-CoA, requires nicotinamide diphosphate (NAD) and Co-enzyme A (CoA) as co-factors. Both of these are components of the premix and hence added to the system to enhance flux through the reaction. The acetyl CoA is phosphorylated by inorganic phosphate, and then de-phosphorylated to produce ATP from ADP. The ATP is used as energy to drive translation of mRNA. |
| − | </br></br> | + | </br> |
| + | </p> | ||
| + | |||
| + | <div> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/9/97/T--Newcastle--BB_CFPS_overview.png" width="800px"/> | ||
| + | <p class="legend"><strong>Figure 1</strong> Diagrammatic overview of CFPS supplement roles in transcription and translation.</p> | ||
| + | </div> | ||
| + | |||
| + | <p> | ||
| + | |||
| + | </br> | ||
Energy can also be derived from glutamate in the supplement solution (Jewett, <i>et al</i>., 2008), which is added in the form of magnesium glutamate and potassium glutamate. Glutamate is a metabolite in the tricarboxylic acid cycle, which generates NADH. In whole cells, NADH is used in oxidative phosphorylation to produce ATP. Oxidative phosphorylation relies on membrane bound proteins and proton gradients across a membrane. It has been shown previously that extracts prepared using French Press or sonication contain membrane vesicles which have ATPase activity (Futai, 1974), and that oxidative phosphorylation can be activated in CFPS systems (Jewett, <i>et al</i>., 2008). | Energy can also be derived from glutamate in the supplement solution (Jewett, <i>et al</i>., 2008), which is added in the form of magnesium glutamate and potassium glutamate. Glutamate is a metabolite in the tricarboxylic acid cycle, which generates NADH. In whole cells, NADH is used in oxidative phosphorylation to produce ATP. Oxidative phosphorylation relies on membrane bound proteins and proton gradients across a membrane. It has been shown previously that extracts prepared using French Press or sonication contain membrane vesicles which have ATPase activity (Futai, 1974), and that oxidative phosphorylation can be activated in CFPS systems (Jewett, <i>et al</i>., 2008). | ||
</br></br> | </br></br> | ||
| Line 1,423: | Line 1,430: | ||
<br /> | <br /> | ||
<p> | <p> | ||
| − | The following solutions were prepared in autoclave sterilised MiliQ water and stored at -80oC: 100x magnesium glutamate solution (1.2 M magnesium glutamate), 10x salt solution (1.3 M potassium glutamate, 40 mM sodium oxalate, 10 mM ammonium acetate), 25x NTPS & co-factor mix (37.5 mM spermidine, 30 mM ATP, 21.25 mM GTP, UTP, and CTP, 25 mM putrescine, 8.25 mM nicotinamide diphosphate, 4.25 mg mL-1 <i>E. coli</i> tRNA (Roche), 0.85 mg mL-1 folinic acid, N xX co-enzyme A), 25x sodium pyruvate solution (825 mM sodium pyruvate, pH to 7.3 with potassium hydroxide), unless stated otherwise. A 5x CFPS supplement solution premix (5% v/v nuclease free water, 5% v/v magnesium glutamate solution, 50% v/v salt solution, 20% v/v NTPS & co-factor mix, 20% v/v sodium pyruvate solution, unless stated otherwise) was prepared and stored at -80oC. CFPS activity of systems prepared as above were tested by expression of 1.7 μg pSB1C3-J23100-sfGFP (Figure | + | The following solutions were prepared in autoclave sterilised MiliQ water and stored at -80oC: 100x magnesium glutamate solution (1.2 M magnesium glutamate), 10x salt solution (1.3 M potassium glutamate, 40 mM sodium oxalate, 10 mM ammonium acetate), 25x NTPS & co-factor mix (37.5 mM spermidine, 30 mM ATP, 21.25 mM GTP, UTP, and CTP, 25 mM putrescine, 8.25 mM nicotinamide diphosphate, 4.25 mg mL-1 <i>E. coli</i> tRNA (Roche), 0.85 mg mL-1 folinic acid, N xX co-enzyme A), 25x sodium pyruvate solution (825 mM sodium pyruvate, pH to 7.3 with potassium hydroxide), unless stated otherwise. A 5x CFPS supplement solution premix (5% v/v nuclease free water, 5% v/v magnesium glutamate solution, 50% v/v salt solution, 20% v/v NTPS & co-factor mix, 20% v/v sodium pyruvate solution, unless stated otherwise) was prepared and stored at -80oC. CFPS activity of systems prepared as above were tested by expression of 1.7 μg pSB1C3-J23100-sfGFP (Figure 2). Firstly, enough CFPS master mix was prepared for 7 reactions by mixing 112 μL cell extract, 70 μL CFPS supplement premix, and 21 μL amino acid solution in a 1.5 mL tube and stored on ice. A further six 1.5 mL tubes were put on ice; 21 μL of nuclease free water was added to three tubes, and 1.7 μg pSB1C3-J23100-sfGFP plasmid DNA from the same stock solution was added to the remaining three. Tubes containing DNA were made up to 21 μL with nuclease-free water. CFPS master mix (29 μL) was then added to all tubes, which were vortexed and transferred to a 96-well plate. The plate was incubated in a BMG Labtech Fluostar Optima at 370C for 4.25 hours with fluorescence readings (excitation: 485 nm, emission: 510 nm) every 15 mins. Figure 3 shows that over time, fluorescence intensity increased in systems with DNA encoding for sfGFP compared to systems with no DNA. Hence, the system had CFPS activity.</p> |
| Line 1,433: | Line 1,440: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/1/1d/T--Newcastle--BB_pSB1C3-sfGFP_plasmid_map.png" width="400px"/> | <img src="https://static.igem.org/mediawiki/2017/1/1d/T--Newcastle--BB_pSB1C3-sfGFP_plasmid_map.png" width="400px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 2:</strong> Plasmid map for pSB1C3-sfGFP. Construct is standard biobrick part BBa_ K515105.</p> |
</div> | </div> | ||
| Line 1,441: | Line 1,448: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/e/e3/T--Newcastle--BB_CFPS_initial_test.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/e/e3/T--Newcastle--BB_CFPS_initial_test.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 3:</strong> Negative corrected fluorescence for <i>E. coli</i> BL21 extract-based CFPS systems. Each data point is an average of 3 replicate reactions, and error bars represent +/- standard error.</p> |
</div> | </div> | ||
| Line 1,468: | Line 1,475: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Experimental Procedure 1</h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Experimental Procedure 1</h2> | ||
| − | <p>Cell extracts were prepared from <i>E. coli</i> BL21 cells using sonication. A CFPS supplement premix solution was prepared as above, except the salts were omitted. Separate solutions for each salt were prepared and added to each CFPS reaction according to the main effects screening design. Reactions were performed as above and CFPS activity was measured as fluorescence at each time point minus fluorescence at 15 mins (Figure | + | <p>Cell extracts were prepared from <i>E. coli</i> BL21 cells using sonication. A CFPS supplement premix solution was prepared as above, except the salts were omitted. Separate solutions for each salt were prepared and added to each CFPS reaction according to the main effects screening design. Reactions were performed as above and CFPS activity was measured as fluorescence at each time point minus fluorescence at 15 mins (Figure 4). Endpoint data was then used, along with the JMP software, to build a model predicting the important factors (Figure 5). |
</br></br> | </br></br> | ||
| Line 1,477: | Line 1,484: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/f/fb/T--Newcastle--BB_CFPS_figure3.png" width="400px" style="background-color: white;"/> | <img src="https://static.igem.org/mediawiki/2017/f/fb/T--Newcastle--BB_CFPS_figure3.png" width="400px" style="background-color: white;"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 4:</strong> CFPS activity of CFPS reactions with salt concentrations according to the main effects screening design (table 2) was determined as fluorescence intensity at each time point minus fluorescence intensity at the 15 minute time point. Reaction IDs are described in table 2.</p> |
</div> | </div> | ||
| Line 1,485: | Line 1,492: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/b/b0/T--Newcastle--BB_CFPS_figure4.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/b/b0/T--Newcastle--BB_CFPS_figure4.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 5:</strong> Screening model constructed using JMP showing which factors were closest to significance. Predictions for interactions are unreliable due to forced orthogonality (*). Of the primary factors, magnesium glutamate is the closest to significant, followed by potassium glutamate, sodium oxalate, and ammonium acetate in that order.</p> |
</div> | </div> | ||
| Line 1,498: | Line 1,505: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/5/5f/T--Newcastle--BB_CFPS_figure5.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/5/5f/T--Newcastle--BB_CFPS_figure5.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 6:</strong> Comparison of four surface response designs generated by the JMP software using the compare designs feature. From top to bottom: design type, number of reactions required by the design, colour map on correlations, Power analysis of terms, efficiencies, and average prediction variance. For the colour map on correlations, red is highly correlated and blue is highly un-correlated. </p> |
</div> | </div> | ||
<br /> | <br /> | ||
<br /> | <br /> | ||
| − | <p>The three salts which the screening design determined as being the most important (magnesium glutamate, potassium glutamate, and sodium oxalate) were analysed further. A surface response design was used to help determine optimal concentrations for each salt in the CFPS supplement premix solution. The JMP software was used to create a classical surface response design for magnesium glutamate, potassium glutamate, and sodium oxalate. Each factor was given a lower limit of 0.5 times their ‘normal’ concentration, and an upper limit of 1.5 times their ‘normal’ concentration. Four types of surface response designs were constructed and compared (Figure | + | <p>The three salts which the screening design determined as being the most important (magnesium glutamate, potassium glutamate, and sodium oxalate) were analysed further. A surface response design was used to help determine optimal concentrations for each salt in the CFPS supplement premix solution. The JMP software was used to create a classical surface response design for magnesium glutamate, potassium glutamate, and sodium oxalate. Each factor was given a lower limit of 0.5 times their ‘normal’ concentration, and an upper limit of 1.5 times their ‘normal’ concentration. Four types of surface response designs were constructed and compared (Figure 6 - see <a href="https://2017.igem.org/Team:Newcastle/Model#doe">our modelling page</a> for more information). Ultimately the central composite design – orthogonal was chosen. The list of experiments determined by this design are shown in Table 3.</p> |
</br></br> | </br></br> | ||
| Line 1,512: | Line 1,519: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Experimental Procedure 2</h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Experimental Procedure 2</h2> | ||
| − | <p>Cell extracts were prepared and CFPS reactions performed as before, except the magnesium glutamate, potassium glutamate, and sodium oxalate concentrations were according to the surface response experimental design. Ammonium acetate was kept at the default amount. CFPS activity was measured as fluorescence at each time point minus fluorescence at 15 mins (Figure | + | <p>Cell extracts were prepared and CFPS reactions performed as before, except the magnesium glutamate, potassium glutamate, and sodium oxalate concentrations were according to the surface response experimental design. Ammonium acetate was kept at the default amount. CFPS activity was measured as fluorescence at each time point minus fluorescence at 15 mins (Figure 7). Endpoint data was then used, along with the JMP software, to build a model predicting optimal concentrations for the three salts analysed (predictions visualised in figure 8). These predictions were then tested by preparing a supplement solution premix with amounts of magnesium glutamate, potassium glutamate, and sodium oxalate at concentrations of 6 mM, 195 mM, and 2 mM respectively. This supplement solution premix was used to supplement two batches of cell extract which were prepared identically. The first batch was the same extract used to collect data on which the predictions were made, whereas the second batch was newly prepared. It was found that for the first extract, CFPS activity was enhanced when the premix containing ‘optimised’ concentrations of salts was used compared to the un-altered supplement solution premix (Figure 9a). Additionally, CFPS activity was observed as being within the confidence intervals predicted by the DoE model. |
</br></br> | </br></br> | ||
| − | When CFPS reactions were performed using the second extract and the new premix, CFPS activity was lower compared to reactions using the second extract and the original premix (Figure | + | When CFPS reactions were performed using the second extract and the new premix, CFPS activity was lower compared to reactions using the second extract and the original premix (Figure 9b). This supports the view that each extract requires separate optimisation of the supplement solution premix to reach optimal CFPS activity, and that a DoE approach is suitable for achieving this. |
</br></br> | </br></br> | ||
</p> | </p> | ||
| Line 1,526: | Line 1,533: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/2/24/T--Newcastle--BB_CFPS_figure6.png" width="400px"/> | <img src="https://static.igem.org/mediawiki/2017/2/24/T--Newcastle--BB_CFPS_figure6.png" width="400px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 7:</strong> CFPS activity of CFPS reactions with salt concentrations according to the surface response design (Table 3) was determined as fluorescence intensity at each time point minus fluorescence intensity at the 15 minute time point. Reactions 12 through 16 failed due to an error in set-up. These reactions were replicates of reactions 8-11 and their removal had little negative effect on the SRD according to the design diagnostics tool in JMP.</p> |
</div> | </div> | ||
| Line 1,534: | Line 1,541: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/4/41/T--Newcastle--BB_CFPS_figure7.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/4/41/T--Newcastle--BB_CFPS_figure7.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 8:</strong> Cube plot generated by JMP using data collected for the SRD. The x-axis shows magnesium glutamate concentration, the y-axis shows potassium glutamate concentration, and the z-axis shows sodium oxalate concentration. The ovals shows predicted CFPS concentration. The maximal CFPS activity was found at 195 mM potassium glutamate, 6 mM magnesium glutamate, and 2 mM sodium oxalate. The minimal point was found at 65 mM potassium glutamate, 18 mM magnesium glutamate, and 2 mM sodium oxalate. This was verified as the maximum and minimum points using the surface profiler function in JMP.</p> |
</div> | </div> | ||
| Line 1,543: | Line 1,550: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/9/99/T--Newcastle--BB_CFPS_figure8.png" width="600px" style="background-color:white;"/> | <img src="https://static.igem.org/mediawiki/2017/9/99/T--Newcastle--BB_CFPS_figure8.png" width="600px" style="background-color:white;"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 9:</strong> CFPS activity for two CFPS systems utilising two different cell extract batches prepared identically. A) Results for a system utilising the same extract batch used to test the SRD. B) Results for a system utilising a new batch of cell extract. The blue lines show systems using the normal CFPS supplement premix, and the purple lines show the systems with supplement premix with ‘optimised’ magnesium glutamate, potassium glutamate, and sodium oxalate as identified above. The system utilising extract from the same batch used in the SRD testing had a higher CFPS activity with the ‘optimised’ premix (purple) than with the original premix (blue), whereas the system utilising extract from a separate batch had higher activity with the original premix.</p> |
</div> | </div> | ||
| Line 1,577: | Line 1,584: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/8/83/T--Newcastle--BB_CFPS_figure9.png" width="400px"/> | <img src="https://static.igem.org/mediawiki/2017/8/83/T--Newcastle--BB_CFPS_figure9.png" width="400px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 10:</strong> Extract batch 1: CFPS activity (fluorescence at each time point minus fluorescence at 15 mins) of reactions with supplements in amounts according to the DoE supplement screening design. Legend shows the reaction number for each system (Table 4).</p> |
</div> | </div> | ||
| Line 1,585: | Line 1,592: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/9/90/T--Newcastle--BB_CFPS_figure10.png" width="600px"/> | <img src="https://static.igem.org/mediawiki/2017/9/90/T--Newcastle--BB_CFPS_figure10.png" width="600px"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 11:</strong> Extract batch 2: CFPS activity (fluorescence at each time point minus fluorescence at 15 mins) of reactions with supplements in amounts according to the DoE supplement screening design. Legend shows the reaction number for each system (Table 4).</p> |
</div> | </div> | ||
| Line 1,595: | Line 1,602: | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2017/5/58/T--Newcastle--BB_CFPS_figure11.png" width="600px" style="background-color:white;"/> | <img src="https://static.igem.org/mediawiki/2017/5/58/T--Newcastle--BB_CFPS_figure11.png" width="600px" style="background-color:white;"/> | ||
| − | <p class="legend"><strong>Figure | + | <p class="legend"><strong>Figure 12:</strong> Bar chart of contrast values for each term in the CFPS supplement solution screening design generated by the JMP software. Contrast values are used as an estimate of a factor’s effect on the response. a) From CFPS system data utilising the moderately active extract (extract 1). b) From the CFPS system data utilising the low activity extract (extract 2).</p> |
</div> | </div> | ||
Revision as of 18:51, 31 October 2017
spacefill
spacefill
Our Experimental Results
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project
































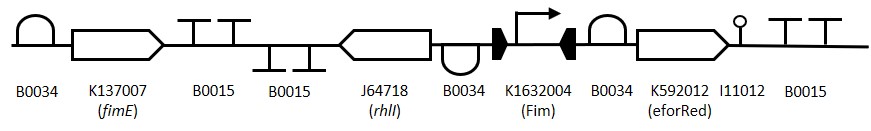
 Figure 2:
Representation of the switching mechanism of the Fim Switch, in the native [OFF] state the eforRED reporter is expressed (shown in red) allowing direct visualisation of the cells. Following the inversion of the promoter region (
Figure 2:
Representation of the switching mechanism of the Fim Switch, in the native [OFF] state the eforRED reporter is expressed (shown in red) allowing direct visualisation of the cells. Following the inversion of the promoter region (
 Figure 3:
High fidelity amplification of the 3 gBlock fragments for assembly of the Fim Switch. The gBlock-1 amplification is shown in lanes 1+2 (819 bp), gBlock-2 amplification is shown in lanes 3+4 (1148 bp) and the gBlock-3 amplification is shown in lanes 5+6 (939bp).
Figure 3:
High fidelity amplification of the 3 gBlock fragments for assembly of the Fim Switch. The gBlock-1 amplification is shown in lanes 1+2 (819 bp), gBlock-2 amplification is shown in lanes 3+4 (1148 bp) and the gBlock-3 amplification is shown in lanes 5+6 (939bp).
 Figure 4:
Patches of the Fim Switch transformants. Patch number 6 shows the correct red colour which indicates expression of the eforRed chromoprotein.
Figure 4:
Patches of the Fim Switch transformants. Patch number 6 shows the correct red colour which indicates expression of the eforRed chromoprotein.













 The
The