| Line 1,218: | Line 1,218: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| − | <p> Biosensors, synthetic systems designed to detect and respond to a target analyte, are a common application of synthetic biology. However, the production and screening of multiple biosensor system variants is hindered by the inefficiency and specificity of the gene assembly techniques used. The production of circuit variants is important in biosensor production, as sensitivity to target molecules must be tuned. Aim: To develop a multicellular biosensor development platform which utilises cell-mixing, as opposed to genetic re-engineering, to construct biosensor variants.</p> | + | <p> Biosensors, synthetic systems designed to detect and respond to a target analyte, are a common application of synthetic biology. However, the production and screening of multiple biosensor system variants is hindered by the inefficiency and specificity of the gene assembly techniques used. The production of circuit variants is important in biosensor production, as sensitivity to target molecules must be tuned. </br></p> |
| + | <p><b>Aim:</b> To develop a multicellular biosensor development platform which utilises cell-mixing, as opposed to genetic re-engineering, to construct biosensor variants.</p> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
| − | <p><b>Human Practices Quotes:</br><p> | + | <p><b>Human Practices Quotes:</br></br> |
| − | "Modularity would be a really useful aspect" - Dr Karen Polizzi </br> | + | <p> |
| − | + | "<i>Modularity would be a really useful aspect</i>" - Dr Karen Polizzi, Imperial college - London </br></br> | |
| − | </b></p> | + | “<i>Each generation of scientists keep reinventing the wheel</i>” - Dr Martin Peacock, |
| + | </b></br></br></p> | ||
<p><b>Biosensor Development.</b> When developing biosensors, it would be useful to test multiple variants of a circuit. This is especially important in the fine-tuning of biosensor behaviour as this requires the screening of many variants to find appropriate activation thresholds for a system. Apart from the initial detection unit, many constructs used in synthetic biology based biosensors are reusable between different biosensor systems, such as fluorescent protein coding sequences or devices which amplify signals. However, these parts rarely get reused. For example, the Cambridge iGEM team (2009) developed a library of sensitivity tuners which were able to convert polymerase per second inputs to a desired polymerase per second output, allowing a biosensor developer control over the sensitivity of their systems to various target analyte concentrations. This project was impressive enough to win the competition. However, despite the parts' clear usefulness, there is no documentation that the parts have ever been successfully reused within the iGEM competition. We suggest that this is due to the difficulties in assembling biosensors systems – the screening of a library of sensitivity tuners would require the ability to easily generate multiple sensor circuits. Although only one part would be changing in each circuit variant, current genetic engineering techniques mean that parts are tightly coupled together, preventing the simple swapping of parts. | <p><b>Biosensor Development.</b> When developing biosensors, it would be useful to test multiple variants of a circuit. This is especially important in the fine-tuning of biosensor behaviour as this requires the screening of many variants to find appropriate activation thresholds for a system. Apart from the initial detection unit, many constructs used in synthetic biology based biosensors are reusable between different biosensor systems, such as fluorescent protein coding sequences or devices which amplify signals. However, these parts rarely get reused. For example, the Cambridge iGEM team (2009) developed a library of sensitivity tuners which were able to convert polymerase per second inputs to a desired polymerase per second output, allowing a biosensor developer control over the sensitivity of their systems to various target analyte concentrations. This project was impressive enough to win the competition. However, despite the parts' clear usefulness, there is no documentation that the parts have ever been successfully reused within the iGEM competition. We suggest that this is due to the difficulties in assembling biosensors systems – the screening of a library of sensitivity tuners would require the ability to easily generate multiple sensor circuits. Although only one part would be changing in each circuit variant, current genetic engineering techniques mean that parts are tightly coupled together, preventing the simple swapping of parts. | ||
</br></br> | </br></br> | ||
| Line 1,247: | Line 1,249: | ||
<p> | <p> | ||
<center><b>Figure 2:</b> Modular and multicellular Sensynova framework design.</center> | <center><b>Figure 2:</b> Modular and multicellular Sensynova framework design.</center> | ||
| − | + | </br></p> | |
<p> The splitting of biosensor components into separate cells may have additional advantages besides ease of variant production. Goni-Moreno <i>et al</i>. (2011) have previously suggested that the use of synthetic quorum sensing circuits enables each cell to be considered an independent logic gate, which may rectify the “fuzzy logic” seen in some biosensors, where stochastic cellular processes may produce false positive results. Quorum sensing has also been previously used to synchronise gene expressions, leading to reduced variability within a population (Danino <i>et al</i>., 2010).</p> | <p> The splitting of biosensor components into separate cells may have additional advantages besides ease of variant production. Goni-Moreno <i>et al</i>. (2011) have previously suggested that the use of synthetic quorum sensing circuits enables each cell to be considered an independent logic gate, which may rectify the “fuzzy logic” seen in some biosensors, where stochastic cellular processes may produce false positive results. Quorum sensing has also been previously used to synchronise gene expressions, leading to reduced variability within a population (Danino <i>et al</i>., 2010).</p> | ||
| Line 1,258: | Line 1,260: | ||
<p>To modularise biosensor components, it was necessary to first confirm which devices types are commonly found in biosensors. An in depth systematic review was conducted to determine these components. Team seeker, a tool for keyword searches of iGEM team titles and abstracts for the years 2008 to 2016, was used to identify biosensor based projects (Aalto-Helsinki iGEM team, 2014). The search terms used to identify potentially relevant projects were “sense” and “biosensor”. 121 projects were identified by these search terms. In projects including multiple sensors, the most well characterised sensors were used for this review. Sensor designs, rather than constructed biosensors, were used for analysis, as time constraints in iGEM often prevents project completion. | <p>To modularise biosensor components, it was necessary to first confirm which devices types are commonly found in biosensors. An in depth systematic review was conducted to determine these components. Team seeker, a tool for keyword searches of iGEM team titles and abstracts for the years 2008 to 2016, was used to identify biosensor based projects (Aalto-Helsinki iGEM team, 2014). The search terms used to identify potentially relevant projects were “sense” and “biosensor”. 121 projects were identified by these search terms. In projects including multiple sensors, the most well characterised sensors were used for this review. Sensor designs, rather than constructed biosensors, were used for analysis, as time constraints in iGEM often prevents project completion. | ||
</br></br> | </br></br> | ||
| − | Ten projects were unable to be reviewed because their wiki was broken. Of the remaining 111 projects, 18 projects were deemed not eligible for further analysis. This was either due to a lack of information regarding biosensor mechanism provided by the team or their project was irrelevant. | + | Ten projects were unable to be reviewed because their wiki was broken. Of the remaining 111 projects, 18 projects were deemed not eligible for further analysis. This was either due to a lack of information regarding biosensor mechanism provided by the team or their project was irrelevant. Three projects were excluded as the sensing component of their project was unchanged from a previous project, to prevent the overrepresentation of biosensors in our database. Therefore, a total of 93 biosensors were used for analysis in our systematic review (Figure 3 and Table 1). </br></br>The systematic review revealed that all biosensors could be split into four components: |
</br> | </br> | ||
<b> 1) Detector: </b>The part responsible for detection of the target molecules. For example, riboswitches and transcription factors. </br> | <b> 1) Detector: </b>The part responsible for detection of the target molecules. For example, riboswitches and transcription factors. </br> | ||
| Line 1,293: | Line 1,295: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | ||
| − | <p>The Framework characterisation has been performed using a BMG-Labtech fluostar optima plate reader in order to monitor the absorbance ( | + | <p>The Framework characterisation has been performed using a BMG-Labtech fluostar optima plate reader in order to monitor the absorbance (OD<sub>600</sub> nm) and GFP fluorescence (excitation 485 nm, emission 510 nm).</p> |
<p> | <p> | ||
| − | The 3 cultures (IPTG detector, processor and sfGFP reporter) were grown separately in LB+ chloramphenicol on a shaker at 37C. After the overnight incubation the cultures were diluted to | + | The 3 cultures (IPTG detector, processor and sfGFP reporter) were grown separately in LB+ chloramphenicol on a shaker at 37C. After the overnight incubation the cultures were diluted to OD<sub>600</sub> 0.1 in order to achieve the syncronised growth to reach the late exponential phase. At OD<sub>600</sub> between 0.5 and 0.7 the cultures were mixed in ratio1:1:1 and IPTG 1mM was added. To test the functionality of the processor and the output, tests with the specific quorum sensing molecules were performed as described below.</p> |
| − | + | </br> | |
<p> | <p> | ||
<b>Reporter test.</b> The culture carrying the reporter device <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> was also tested individually after induction with the connector C12-RHL 2 ug/ul as shown in the graph (Figure 6). </p> | <b>Reporter test.</b> The culture carrying the reporter device <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> was also tested individually after induction with the connector C12-RHL 2 ug/ul as shown in the graph (Figure 6). </p> | ||
| − | + | </br> | |
<img src="https://static.igem.org/mediawiki/parts/1/19/Rhl_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | <img src="https://static.igem.org/mediawiki/parts/1/19/Rhl_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | ||
<p> | <p> | ||
| Line 1,310: | Line 1,312: | ||
<p> | <p> | ||
| − | <b> Processor test.</b> The connector 1 (C4-HSL) was added to the co-culture consisting of processor <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> + reporter <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> in ratio 1:1. The plot shows the successful communication via quorum sensing in the Sensynova device. It is clear that the presence of 1mM C4-HSL is detected by the processor cells which produce the connector 2 (C12-RHL) for the reporter cells to detect. This induction in the reporter cells leads to the expression of sfGFP (Figure 7).</p> | + | <b> Processor test.</b> The connector 1 (C4-HSL) was added to the co-culture consisting of processor <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> + reporter <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> in ratio 1:1. The plot shows the successful communication via quorum sensing in the Sensynova device. It is clear that the presence of 1mM C4-HSL is detected by the processor cells which produce the connector 2 (C12-RHL) for the reporter cells to detect. This induction in the reporter cells leads to the expression of sfGFP (Figure 7).</p></br> |
<img src="https://static.igem.org/mediawiki/parts/c/c6/Pro_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | <img src="https://static.igem.org/mediawiki/parts/c/c6/Pro_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | ||
| Line 1,320: | Line 1,322: | ||
<p> | <p> | ||
| − | <b> Framework test.</b> The co-culture of the 3 cell types was inoculated at ratio 1:1:1 (detectors:processors:reporters), growth and fluorescence were monitored after induction with IPTG 1mM. The plot shows no significant increasing fluorescence in the induced samples (Figure 8).</p> | + | <b> Framework test.</b> The co-culture of the 3 cell types was inoculated at ratio 1:1:1 (detectors:processors:reporters), growth and fluorescence were monitored after induction with IPTG 1mM. The plot shows no significant increasing fluorescence in the induced samples (Figure 8).</p></br> |
<img src="https://static.igem.org/mediawiki/parts/f/f2/T--Newcastle--BB_framework_framework_green.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | <img src="https://static.igem.org/mediawiki/parts/f/f2/T--Newcastle--BB_framework_framework_green.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | ||
| Line 1,329: | Line 1,331: | ||
</center></p> </br> | </center></p> </br> | ||
<p> | <p> | ||
| − | Results from the <a href="https://2017.igem.org/Team:Newcastle/Model#sim">multicellular modelling</a> predicted that the traditionally used 1:1:1 ratio is not the optimal combination for the Sensynova device to work. It is in fact suggested to adopt a higher concentration of the reporter culture compare with the detector and processor. Thus, the framework test was repeated incorporating our in silico simulation data and combining the 3 cell types in ratio 1:1:13 (detectors:processors:reporters). The experiment results, shown in the picture below, confirm the modelling data. There is a consistent discrepancy between IPTG induced and non-induced samples in the 1:1:13 co-cultures, in comparison with the 1:1:1 co-cultures which don't show any difference in presence or absence of IPTG ( | + | Results from the <a href="https://2017.igem.org/Team:Newcastle/Model#sim">multicellular modelling</a> predicted that the traditionally used 1:1:1 ratio is not the optimal combination for the Sensynova device to work. It is in fact suggested to adopt a higher concentration of the reporter culture compare with the detector and processor. Thus, the framework test was repeated incorporating our in silico simulation data and combining the 3 cell types in ratio 1:1:13 (detectors:processors:reporters). The experiment results, shown in the picture below, confirm the modelling data. There is a consistent discrepancy between IPTG induced and non-induced samples in the 1:1:13 co-cultures, in comparison with the 1:1:1 co-cultures which don't show any difference in presence or absence of IPTG (Figure 9).</p></br> |
<p> | <p> | ||
<img src="https://static.igem.org/mediawiki/2017/0/0d/T--Newcastle--BB_framework_framework_green1_1_13.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | <img src="https://static.igem.org/mediawiki/2017/0/0d/T--Newcastle--BB_framework_framework_green1_1_13.jpg" class="img-fluid rounded mx-auto d-block" style="max-width: 60%" alt=""> | ||
| Line 1,338: | Line 1,340: | ||
<p> | <p> | ||
<p class="legend"><center><strong><b>Figure 9:</b></strong> Framework (<a href="http://parts.igem.org/Part:BBa_K2205009">BBa_K2205009</a> , <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> , <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> ) test with a co-culture in ratio 1:1:13 in response of IPTG induction. | <p class="legend"><center><strong><b>Figure 9:</b></strong> Framework (<a href="http://parts.igem.org/Part:BBa_K2205009">BBa_K2205009</a> , <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> , <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> ) test with a co-culture in ratio 1:1:13 in response of IPTG induction. | ||
| − | </center></p> | + | </center></p></br> |
<p> | <p> | ||
The experimental data validate the model prediction showing that the system worked most optimally when the reporter cells were in excess of both the detector and processor cells. One of the reasons that this configuration was the best may be because of signal amplification at each of the quorum sensing communication stages. The quorum sensing mechanism used here is the acyl homoserine lactone (AHL) system in gram negative bacteria. This system works by one cell producing a quorum sensing molecule which can diffuse out through its membrane. Once the extracellular space reaches a certain threshold concentration of AHL molecule, the AHL will begin to diffuse into other cells in the community. If the cell the AHL molecule enters has the appropriate transcription factor present (e.g. LasR for the C12 AHL), then transcription of a gene under the control of the pLas promoter can occur. Therefore, if background expression of the AHL is high enough to reach above the threshold level, then expression of the next quorum sensing molecule in another cell (in this case C4 AHL) will occur. By reducing the amount of detector and processor cells present in the system, the background expression levels of C12 and C4 will be lower, and hence expression of sfGFP by the reporter cell will be lower. | The experimental data validate the model prediction showing that the system worked most optimally when the reporter cells were in excess of both the detector and processor cells. One of the reasons that this configuration was the best may be because of signal amplification at each of the quorum sensing communication stages. The quorum sensing mechanism used here is the acyl homoserine lactone (AHL) system in gram negative bacteria. This system works by one cell producing a quorum sensing molecule which can diffuse out through its membrane. Once the extracellular space reaches a certain threshold concentration of AHL molecule, the AHL will begin to diffuse into other cells in the community. If the cell the AHL molecule enters has the appropriate transcription factor present (e.g. LasR for the C12 AHL), then transcription of a gene under the control of the pLas promoter can occur. Therefore, if background expression of the AHL is high enough to reach above the threshold level, then expression of the next quorum sensing molecule in another cell (in this case C4 AHL) will occur. By reducing the amount of detector and processor cells present in the system, the background expression levels of C12 and C4 will be lower, and hence expression of sfGFP by the reporter cell will be lower. | ||
<p> | <p> | ||
</p> </br> | </p> </br> | ||
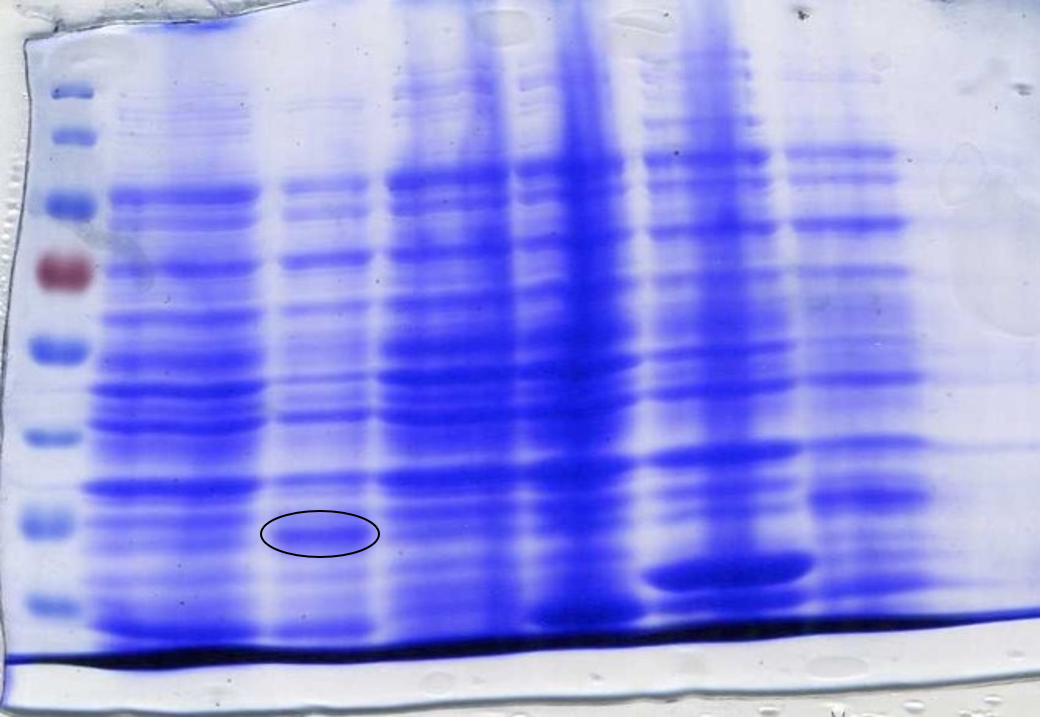
| − | <p><b> Qualitative test with chromoproteins expression. </b>In order to check the performance of the Sensynova device in terms of modularity, cultures of IPTG detector, processor unit and 3 different reporter modules carrying 2 chromoproteins (Chromoproteins link)(<a href="http://parts.igem.org/Part:BBa_K2205016">BBa_K2205016</a>, <a href="http://parts.igem.org/Part:BBa_K2205018">BBa_K2205018</a>)and sfGFP(<a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a>) were inoculated and grown overnight in LB+ | + | <p><b> Qualitative test with chromoproteins expression. </b>In order to check the performance of the Sensynova device in terms of modularity, cultures of IPTG detector, processor unit and 3 different reporter modules carrying 2 chromoproteins (Chromoproteins link)(<a href="http://parts.igem.org/Part:BBa_K2205016">BBa_K2205016</a>, <a href="http://parts.igem.org/Part:BBa_K2205018">BBa_K2205018</a>) and sfGFP (<a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a>) were inoculated and grown overnight in LB+Chloramphenicol (12,5ng/ul). The day after the cultures were diluted at OD600: 0,1 and mixed together to obtain co-cultures with ratio 1:1:1 and 1:1:13. Some samples were supplemented with 1mM IPTG to induce the expression of quorum sensing molecules and eventually achieve the chromoproteins visualisation (Figures 10, 11, 12). |
Revision as of 20:59, 31 October 2017
spacefill
spacefill
Our Experimental Results
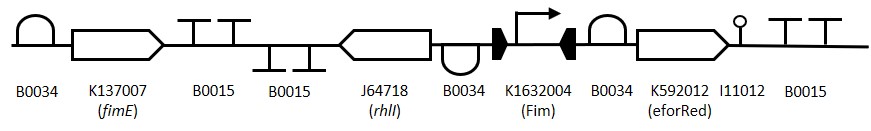
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project





















































 The
The