| Line 1,237: | Line 1,237: | ||
Therefore, we propose a modular, multicellular system for biosensor development, using a cell-to-cell communication system to eradicate the requirement for further genetic engineering of reusable biosensor devices (Figure 1). | Therefore, we propose a modular, multicellular system for biosensor development, using a cell-to-cell communication system to eradicate the requirement for further genetic engineering of reusable biosensor devices (Figure 1). | ||
</br></br> | </br></br> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/7/75/T--Newcastle--BB_biosensor_modules_abstract.png" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/7/75/T--Newcastle--BB_biosensor_modules_abstract.png" class="img-fluid rounded mx-auto d-block" alt=""> |
| − | <img src="https://static.igem.org/mediawiki/2017/0/03/T--Newcastle--BB_framework_framework.png" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/0/03/T--Newcastle--BB_framework_framework.png" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
<center><b>Figure 1: </b> Multicellular Sensynova system.</center> | <center><b>Figure 1: </b> Multicellular Sensynova system.</center> | ||
| Line 1,252: | Line 1,252: | ||
</br></br> | </br></br> | ||
| − | <img src="https://static.igem.org/mediawiki/parts/6/63/Framework_generic.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/parts/6/63/Framework_generic.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
<center><b>Figure 2:</b> <a href="http://sbolstandard.org/visual#post-780">SBOL Visual</a> for the modular and multicellular Sensynova framework design.</center> | <center><b>Figure 2:</b> <a href="http://sbolstandard.org/visual#post-780">SBOL Visual</a> for the modular and multicellular Sensynova framework design.</center> | ||
| Line 1,273: | Line 1,273: | ||
<b>4) Adaptors: </b> If the molecule is hard to detect, adaptor components can be placed before the detector unit, to convert the target molecules to something able to be sensed by the detector component. For example, for target that degrades into an easily detectable molecule, a biochemical conversion adaptor could be placed before the detector component which enzymatically degrades the target molecule into the molecule detected by the detector module. | <b>4) Adaptors: </b> If the molecule is hard to detect, adaptor components can be placed before the detector unit, to convert the target molecules to something able to be sensed by the detector component. For example, for target that degrades into an easily detectable molecule, a biochemical conversion adaptor could be placed before the detector component which enzymatically degrades the target molecule into the molecule detected by the detector module. | ||
</br></br></br> | </br></br></br> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/5/5e/Graphy%281%29.jpeg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/5/5e/Graphy%281%29.jpeg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
<center><b>Figure 3:</b> Frequency of projects based on biosensors development in iGEM. </center></p> | <center><b>Figure 3:</b> Frequency of projects based on biosensors development in iGEM. </center></p> | ||
| Line 1,279: | Line 1,279: | ||
<p> | <p> | ||
<center><b>Table 1: </b>Percentages of biosensors components used in iGEM. </center></p> | <center><b>Table 1: </b>Percentages of biosensors components used in iGEM. </center></p> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/2/25/Igembiosensors_table.png" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/2/25/Igembiosensors_table.png" class="img-fluid rounded mx-auto d-block" alt=""> |
</br></br> | </br></br> | ||
| Line 1,287: | Line 1,287: | ||
<p>To prove that our concept of splitting biosensors across multiple cells would work, we designed an IPTG sensor. The design of this system can be found in Figure 4. In this system, LacI is constitutively expressed in the detector cell and represses the production of LasI. When IPTG is added, it binds LacI, preventing repression. Therefore, in the presence of IPTG, LasI will produce C12, our first connector molecule. To determine that our system would work, it was first tested in silico. Details on the model of this system can be found on our <a href="https://2017.igem.org/Team:Newcastle/Model#sim">Modelling page</a>. | <p>To prove that our concept of splitting biosensors across multiple cells would work, we designed an IPTG sensor. The design of this system can be found in Figure 4. In this system, LacI is constitutively expressed in the detector cell and represses the production of LasI. When IPTG is added, it binds LacI, preventing repression. Therefore, in the presence of IPTG, LasI will produce C12, our first connector molecule. To determine that our system would work, it was first tested in silico. Details on the model of this system can be found on our <a href="https://2017.igem.org/Team:Newcastle/Model#sim">Modelling page</a>. | ||
</br></br> | </br></br> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/5/5c/Iptg_framework.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/5/5c/Iptg_framework.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
<center><b>Figure 4:</b> <a href="http://sbolstandard.org/visual#post-780">SBOL Visual</a> for the Sensynova framework design used for sensing IPTG. </center></p> | <center><b>Figure 4:</b> <a href="http://sbolstandard.org/visual#post-780">SBOL Visual</a> for the Sensynova framework design used for sensing IPTG. </center></p> | ||
| Line 1,294: | Line 1,294: | ||
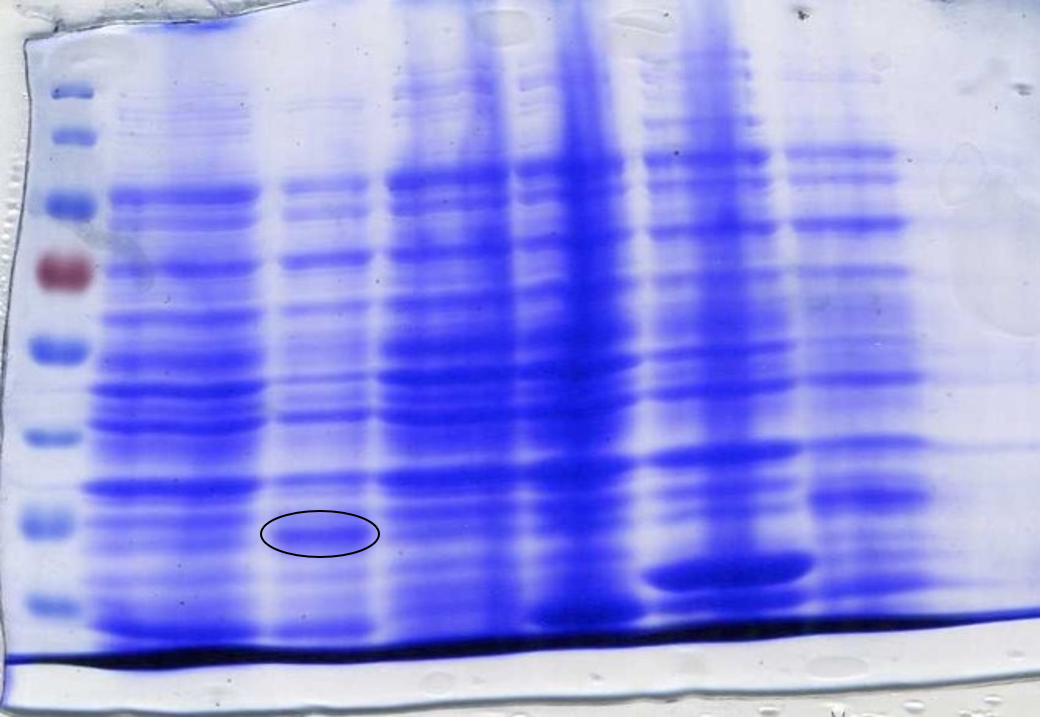
<p>Parts were synthesised by IDT and integration into the pSB1C3 plasmid confirmed by colony PCR and subsequent sequencing. Red boxes show parts later used for biobrick production (Figure 5). </p></br> | <p>Parts were synthesised by IDT and integration into the pSB1C3 plasmid confirmed by colony PCR and subsequent sequencing. Red boxes show parts later used for biobrick production (Figure 5). </p></br> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/1/10/Framework_gel_parts.png" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/1/10/Framework_gel_parts.png" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
<center><b>Figure 5:</b> Colony PCR confirming the correct integration of the biosensor fragments into the vector. </center></p> | <center><b>Figure 5:</b> Colony PCR confirming the correct integration of the biosensor fragments into the vector. </center></p> | ||
| Line 1,308: | Line 1,308: | ||
<b>Reporter test.</b> The culture carrying the reporter device <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> was also tested individually after induction with the connector C12-RHL 2 ug/ul as shown in the graph (Figure 6). </p> | <b>Reporter test.</b> The culture carrying the reporter device <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> was also tested individually after induction with the connector C12-RHL 2 ug/ul as shown in the graph (Figure 6). </p> | ||
</br> | </br> | ||
| − | <img src="https://static.igem.org/mediawiki/parts/1/19/Rhl_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/parts/1/19/Rhl_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/9/97/Rhl_reporter.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/9/97/Rhl_reporter.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
| Line 1,319: | Line 1,319: | ||
<p> | <p> | ||
<b> Processor test.</b> The connector 1 (C4-HSL) was added to the co-culture consisting of processor <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> + reporter <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> in ratio 1:1. The plot shows the successful communication via quorum sensing in the Sensynova device. It is clear that the presence of 1mM C4-HSL is detected by the processor cells which produce the connector 2 (C12-RHL) for the reporter cells to detect. This induction in the reporter cells leads to the expression of sfGFP (Figure 7).</p></br> | <b> Processor test.</b> The connector 1 (C4-HSL) was added to the co-culture consisting of processor <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> + reporter <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> in ratio 1:1. The plot shows the successful communication via quorum sensing in the Sensynova device. It is clear that the presence of 1mM C4-HSL is detected by the processor cells which produce the connector 2 (C12-RHL) for the reporter cells to detect. This induction in the reporter cells leads to the expression of sfGFP (Figure 7).</p></br> | ||
| − | <img src="https://static.igem.org/mediawiki/parts/c/c6/Pro_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/parts/c/c6/Pro_rep_fluo.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
| − | <img src="https://static.igem.org/mediawiki/2017/f/f0/Hsl_proc_rep.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/f/f0/Hsl_proc_rep.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p><center> <b>Figure 7:</b> The processor <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> and reporter <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> co-culture test shows increasing fluorescence/time after specific induction with the connecting molecule C12-RHL. | <p><center> <b>Figure 7:</b> The processor <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> and reporter <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> co-culture test shows increasing fluorescence/time after specific induction with the connecting molecule C12-RHL. | ||
| Line 1,329: | Line 1,329: | ||
<b> Framework test.</b> The co-culture of the 3 cell types was inoculated at ratio 1:1:1 (detectors:processors:reporters), growth and fluorescence were monitored after induction with IPTG 1mM. The plot shows no significant increasing fluorescence in the induced samples (Figure 8).</p></br> | <b> Framework test.</b> The co-culture of the 3 cell types was inoculated at ratio 1:1:1 (detectors:processors:reporters), growth and fluorescence were monitored after induction with IPTG 1mM. The plot shows no significant increasing fluorescence in the induced samples (Figure 8).</p></br> | ||
| − | <img src="https://static.igem.org/mediawiki/parts/f/f2/T--Newcastle--BB_framework_framework_green.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/parts/f/f2/T--Newcastle--BB_framework_framework_green.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
| − | <img src="https://static.igem.org/mediawiki/parts/0/0d/T--Newcastle--BB_framework_framework_green1_1_13.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/parts/0/0d/T--Newcastle--BB_framework_framework_green1_1_13.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
</p> | </p> | ||
<p><center> <b>Figure 8:</b> Framework (<a href="http://parts.igem.org/Part:BBa_K2205009">BBa_K2205009</a> , <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> , <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> ) test with a co-culture in ratio 1:1:1 in response of IPTG induction. | <p><center> <b>Figure 8:</b> Framework (<a href="http://parts.igem.org/Part:BBa_K2205009">BBa_K2205009</a> , <a href="http://parts.igem.org/Part:BBa_K2205012">BBa_K2205012</a> , <a href="http://parts.igem.org/Part:BBa_K2205015">BBa_K2205015</a> ) test with a co-culture in ratio 1:1:1 in response of IPTG induction. | ||
| Line 1,339: | Line 1,339: | ||
Results from the <a href="https://2017.igem.org/Team:Newcastle/Model#sim">multicellular modelling</a> predicted that the traditionally used 1:1:1 ratio is not the optimal combination for the Sensynova device to work. It is in fact suggested to adopt a higher concentration of the reporter culture compare with the detector and processor. Thus, the framework test was repeated incorporating our in silico simulation data and combining the 3 cell types in ratio 1:1:13 (detectors:processors:reporters). The experiment results, shown in the picture below, confirm the modelling data. There is a consistent discrepancy between IPTG induced and non-induced samples in the 1:1:13 co-cultures, in comparison with the 1:1:1 co-cultures which don't show any difference in presence or absence of IPTG (Figure 9).</p></br> | Results from the <a href="https://2017.igem.org/Team:Newcastle/Model#sim">multicellular modelling</a> predicted that the traditionally used 1:1:1 ratio is not the optimal combination for the Sensynova device to work. It is in fact suggested to adopt a higher concentration of the reporter culture compare with the detector and processor. Thus, the framework test was repeated incorporating our in silico simulation data and combining the 3 cell types in ratio 1:1:13 (detectors:processors:reporters). The experiment results, shown in the picture below, confirm the modelling data. There is a consistent discrepancy between IPTG induced and non-induced samples in the 1:1:13 co-cultures, in comparison with the 1:1:1 co-cultures which don't show any difference in presence or absence of IPTG (Figure 9).</p></br> | ||
<p> | <p> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/0/0d/T--Newcastle--BB_framework_framework_green1_1_13.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/2017/0/0d/T--Newcastle--BB_framework_framework_green1_1_13.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
| − | <img src="https://static.igem.org/mediawiki/parts/1/15/Ratios22.jpg" class="img-fluid rounded mx-auto d-block | + | <img src="https://static.igem.org/mediawiki/parts/1/15/Ratios22.jpg" class="img-fluid rounded mx-auto d-block" alt=""> |
<p> | <p> | ||
| Line 1,456: | Line 1,456: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/9/97/T--Newcastle--BB_CFPS_overview.png" width="800px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/9/97/T--Newcastle--BB_CFPS_overview.png" width="800px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 1</strong> Diagrammatic overview of CFPS supplement roles in transcription and translation.</center></p> | <p class="legend"><center><strong>Figure 1</strong> Diagrammatic overview of CFPS supplement roles in transcription and translation.</center></p> | ||
</div> | </div> | ||
| Line 1,486: | Line 1,486: | ||
<br /> | <br /> | ||
<p class="legend"><center><strong>Table 1:</strong> Concentrations for amino acid stock solutions (first column), 10x amino acid mix (second column), and for each CFPS system (third column).</center></p> | <p class="legend"><center><strong>Table 1:</strong> Concentrations for amino acid stock solutions (first column), 10x amino acid mix (second column), and for each CFPS system (third column).</center></p> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/f/fd/T--Newcastle--BB_amino_acid_amounts.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/f/fd/T--Newcastle--BB_amino_acid_amounts.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
</div> | </div> | ||
| Line 1,501: | Line 1,501: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/1/1d/T--Newcastle--BB_pSB1C3-sfGFP_plasmid_map.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%; | + | <img src="https://static.igem.org/mediawiki/2017/1/1d/T--Newcastle--BB_pSB1C3-sfGFP_plasmid_map.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%; alt=""/> |
<p class="legend"><center><strong>Figure 2:</strong> Plasmid map for pSB1C3-sfGFP. Construct is standard biobrick part BBa_ K515105.</center></p> | <p class="legend"><center><strong>Figure 2:</strong> Plasmid map for pSB1C3-sfGFP. Construct is standard biobrick part BBa_ K515105.</center></p> | ||
</div> | </div> | ||
| Line 1,509: | Line 1,509: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/e/e3/T--Newcastle--BB_CFPS_initial_test.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/e/e3/T--Newcastle--BB_CFPS_initial_test.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 3:</strong> Negative corrected fluorescence for <i>E. coli</i> BL21 extract-based CFPS systems. Each data point is an average of 3 replicate reactions, and error bars represent +/- standard error.</center></p> | <p class="legend"><center><strong>Figure 3:</strong> Negative corrected fluorescence for <i>E. coli</i> BL21 extract-based CFPS systems. Each data point is an average of 3 replicate reactions, and error bars represent +/- standard error.</center></p> | ||
</div> | </div> | ||
| Line 1,545: | Line 1,545: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/f/fb/T--Newcastle--BB_CFPS_figure3.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/f/fb/T--Newcastle--BB_CFPS_figure3.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 4:</strong> CFPS activity of CFPS reactions with salt concentrations according to the main effects screening design (table 2) was determined as fluorescence intensity at each time point minus fluorescence intensity at the 15 minute time point. Reaction IDs are described in table 2.</center></p> | <p class="legend"><center><strong>Figure 4:</strong> CFPS activity of CFPS reactions with salt concentrations according to the main effects screening design (table 2) was determined as fluorescence intensity at each time point minus fluorescence intensity at the 15 minute time point. Reaction IDs are described in table 2.</center></p> | ||
</div> | </div> | ||
| Line 1,553: | Line 1,553: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/b/b0/T--Newcastle--BB_CFPS_figure4.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%; | + | <img src="https://static.igem.org/mediawiki/2017/b/b0/T--Newcastle--BB_CFPS_figure4.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%; alt=""/> |
<p class="legend"><center><strong>Figure 5:</strong> Screening model constructed using JMP showing which factors were closest to significance. Predictions for interactions are unreliable due to forced orthogonality (*). Of the primary factors, magnesium glutamate is the closest to significant, followed by potassium glutamate, sodium oxalate, and ammonium acetate in that order.</center></p> | <p class="legend"><center><strong>Figure 5:</strong> Screening model constructed using JMP showing which factors were closest to significance. Predictions for interactions are unreliable due to forced orthogonality (*). Of the primary factors, magnesium glutamate is the closest to significant, followed by potassium glutamate, sodium oxalate, and ammonium acetate in that order.</center></p> | ||
</div> | </div> | ||
| Line 1,566: | Line 1,566: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/5/5f/T--Newcastle--BB_CFPS_figure5.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/5/5f/T--Newcastle--BB_CFPS_figure5.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 6:</strong> Comparison of four surface response designs generated by the JMP software using the compare designs feature. From top to bottom: design type, number of reactions required by the design, colour map on correlations, Power analysis of terms, efficiencies, and average prediction variance. For the colour map on correlations, red is highly correlated and blue is highly un-correlated. </center></p> | <p class="legend"><center><strong>Figure 6:</strong> Comparison of four surface response designs generated by the JMP software using the compare designs feature. From top to bottom: design type, number of reactions required by the design, colour map on correlations, Power analysis of terms, efficiencies, and average prediction variance. For the colour map on correlations, red is highly correlated and blue is highly un-correlated. </center></p> | ||
</div> | </div> | ||
| Line 1,577: | Line 1,577: | ||
<div style="width:310px;"> | <div style="width:310px;"> | ||
<p class="legend"><center><strong>Table 3:</strong> Table of reactions performed according to the DoE salt supplement surface response design. CFPS reactions contained concentrations of magnesium glutamate, potassium glutamate, and sodium oxalate, according to the table above. The pattern column shows how much of each supplement was present in a reaction; very low concentration (a), low concentration (−), usual concentration (0), high concentration (+), and very high concentration (A).</center></p> | <p class="legend"><center><strong>Table 3:</strong> Table of reactions performed according to the DoE salt supplement surface response design. CFPS reactions contained concentrations of magnesium glutamate, potassium glutamate, and sodium oxalate, according to the table above. The pattern column shows how much of each supplement was present in a reaction; very low concentration (a), low concentration (−), usual concentration (0), high concentration (+), and very high concentration (A).</center></p> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/4/4d/T--Newcastle--BB_CFPS_table3.png" width="300px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/4/4d/T--Newcastle--BB_CFPS_table3.png" width="300px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
</div> | </div> | ||
| Line 1,594: | Line 1,594: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/2/24/T--Newcastle--BB_CFPS_figure6.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/2/24/T--Newcastle--BB_CFPS_figure6.png" width="400px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 7:</strong> CFPS activity of CFPS reactions with salt concentrations according to the surface response design (Table 3) was determined as fluorescence intensity at each time point minus fluorescence intensity at the 15 minute time point. Reactions 12 through 16 failed due to an error in set-up. These reactions were replicates of reactions 8-11 and their removal had little negative effect on the SRD according to the design diagnostics tool in JMP.</center></p> | <p class="legend"><center><strong>Figure 7:</strong> CFPS activity of CFPS reactions with salt concentrations according to the surface response design (Table 3) was determined as fluorescence intensity at each time point minus fluorescence intensity at the 15 minute time point. Reactions 12 through 16 failed due to an error in set-up. These reactions were replicates of reactions 8-11 and their removal had little negative effect on the SRD according to the design diagnostics tool in JMP.</center></p> | ||
</div> | </div> | ||
| Line 1,602: | Line 1,602: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/4/41/T--Newcastle--BB_CFPS_figure7.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/4/41/T--Newcastle--BB_CFPS_figure7.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 8:</strong> Cube plot generated by JMP using data collected for the SRD. The x-axis shows magnesium glutamate concentration, the y-axis shows potassium glutamate concentration, and the z-axis shows sodium oxalate concentration. The ovals shows predicted CFPS concentration. The maximal CFPS activity was found at 195 mM potassium glutamate, 6 mM magnesium glutamate, and 2 mM sodium oxalate. The minimal point was found at 65 mM potassium glutamate, 18 mM magnesium glutamate, and 2 mM sodium oxalate. This was verified as the maximum and minimum points using the surface profiler function in JMP.</center></p> | <p class="legend"><center><strong>Figure 8:</strong> Cube plot generated by JMP using data collected for the SRD. The x-axis shows magnesium glutamate concentration, the y-axis shows potassium glutamate concentration, and the z-axis shows sodium oxalate concentration. The ovals shows predicted CFPS concentration. The maximal CFPS activity was found at 195 mM potassium glutamate, 6 mM magnesium glutamate, and 2 mM sodium oxalate. The minimal point was found at 65 mM potassium glutamate, 18 mM magnesium glutamate, and 2 mM sodium oxalate. This was verified as the maximum and minimum points using the surface profiler function in JMP.</center></p> | ||
</div> | </div> | ||
| Line 1,611: | Line 1,611: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/9/99/T--Newcastle--BB_CFPS_figure8.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/9/99/T--Newcastle--BB_CFPS_figure8.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 9:</strong> CFPS activity for two CFPS systems utilising two different cell extract batches prepared identically. A) Results for a system utilising the same extract batch used to test the SRD. B) Results for a system utilising a new batch of cell extract. The blue lines show systems using the normal CFPS supplement premix, and the purple lines show the systems with supplement premix with ‘optimised’ magnesium glutamate, potassium glutamate, and sodium oxalate as identified above. The system utilising extract from the same batch used in the SRD testing had a higher CFPS activity with the ‘optimised’ premix (purple) than with the original premix (blue), whereas the system utilising extract from a separate batch had higher activity with the original premix.</center></p> | <p class="legend"><center><strong>Figure 9:</strong> CFPS activity for two CFPS systems utilising two different cell extract batches prepared identically. A) Results for a system utilising the same extract batch used to test the SRD. B) Results for a system utilising a new batch of cell extract. The blue lines show systems using the normal CFPS supplement premix, and the purple lines show the systems with supplement premix with ‘optimised’ magnesium glutamate, potassium glutamate, and sodium oxalate as identified above. The system utilising extract from the same batch used in the SRD testing had a higher CFPS activity with the ‘optimised’ premix (purple) than with the original premix (blue), whereas the system utilising extract from a separate batch had higher activity with the original premix.</center></p> | ||
</div> | </div> | ||
| Line 1,625: | Line 1,625: | ||
<div> | <div> | ||
<p class="legend"><center><strong>Table 4:</strong> Table of reactions performed according to the DoE full supplement solution screening design. CFPS reactions contained concentrations of each supplement according to the table above.</center></p> | <p class="legend"><center><strong>Table 4:</strong> Table of reactions performed according to the DoE full supplement solution screening design. CFPS reactions contained concentrations of each supplement according to the table above.</center></p> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/e/e2/T--Newcastle--BB_CFPS_Table4.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/e/e2/T--Newcastle--BB_CFPS_Table4.png" width="600px" class="img-fluid border border-dark rounded mx-auto d-block" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt=""/> |
</div> | </div> | ||
| Line 1,645: | Line 1,645: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/8/83/T--Newcastle--BB_CFPS_figure9.png" width="400px" margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/8/83/T--Newcastle--BB_CFPS_figure9.png" width="400px" margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 10:</strong> Extract batch 1: CFPS activity (fluorescence at each time point minus fluorescence at 15 mins) of reactions with supplements in amounts according to the DoE supplement screening design. Legend shows the reaction number for each system (Table 4).</center></p> | <p class="legend"><center><strong>Figure 10:</strong> Extract batch 1: CFPS activity (fluorescence at each time point minus fluorescence at 15 mins) of reactions with supplements in amounts according to the DoE supplement screening design. Legend shows the reaction number for each system (Table 4).</center></p> | ||
</div> | </div> | ||
| Line 1,653: | Line 1,653: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/9/90/T--Newcastle--BB_CFPS_figure10.png" width="600px" margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/9/90/T--Newcastle--BB_CFPS_figure10.png" width="600px" margin-right: 2%; margin-bottom: 2%;" alt=""/> |
<p class="legend"><center><strong>Figure 11:</strong> Extract batch 2: CFPS activity (fluorescence at each time point minus fluorescence at 15 mins) of reactions with supplements in amounts according to the DoE supplement screening design. Legend shows the reaction number for each system (Table 4).</center></p> | <p class="legend"><center><strong>Figure 11:</strong> Extract batch 2: CFPS activity (fluorescence at each time point minus fluorescence at 15 mins) of reactions with supplements in amounts according to the DoE supplement screening design. Legend shows the reaction number for each system (Table 4).</center></p> | ||
</div> | </div> | ||
| Line 1,663: | Line 1,663: | ||
<div> | <div> | ||
| − | <img src="https://static.igem.org/mediawiki/2017/5/58/T--Newcastle--BB_CFPS_figure11.png" width="600px" style="background-color:white; margin-right: 2%; margin-bottom: 2 | + | <img src="https://static.igem.org/mediawiki/2017/5/58/T--Newcastle--BB_CFPS_figure11.png" width="600px" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt="" class="img-fluid border border-dark rounded mx-auto d-block"> |
<p class="legend"><center><strong>Figure 12:</strong> Bar chart of contrast values for each term in the CFPS supplement solution screening design generated by the JMP software. Contrast values are used as an estimate of a factor’s effect on the response. a) From CFPS system data utilising the moderately active extract (extract 1). b) From the CFPS system data utilising the low activity extract (extract 2).</center></p> | <p class="legend"><center><strong>Figure 12:</strong> Bar chart of contrast values for each term in the CFPS supplement solution screening design generated by the JMP software. Contrast values are used as an estimate of a factor’s effect on the response. a) From CFPS system data utilising the moderately active extract (extract 1). b) From the CFPS system data utilising the low activity extract (extract 2).</center></p> | ||
</div> | </div> | ||
Revision as of 21:40, 31 October 2017
spacefill
spacefill
Our Experimental Results
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project






















































 The
The