| Line 966: | Line 966: | ||
</p> | </p> | ||
<h2 style="text-align: left; clear: both"> Future Work </h2> | <h2 style="text-align: left; clear: both"> Future Work </h2> | ||
| + | </br> | ||
<p> | <p> | ||
Since there is some leaky expression of the <i>fimE</i> gene (even without a promoter) to fine tune the Fim switch the lac operator could be inserted upstream of the <i>fimE</i> RBS to repress unwanted expression. The part could then be used following the addition of IPTG. An alternative method would be to clone a transcriptional terminator upstream of the <i>fimE</i> RBS to prevent leaky expression from elsewhere in the plasmid.<br/><br/> | Since there is some leaky expression of the <i>fimE</i> gene (even without a promoter) to fine tune the Fim switch the lac operator could be inserted upstream of the <i>fimE</i> RBS to repress unwanted expression. The part could then be used following the addition of IPTG. An alternative method would be to clone a transcriptional terminator upstream of the <i>fimE</i> RBS to prevent leaky expression from elsewhere in the plasmid.<br/><br/> | ||
| Line 991: | Line 992: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| + | </br> | ||
<p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper the success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. | <p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper the success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. | ||
</br></br> | </br></br> | ||
| Line 997: | Line 999: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
| + | </br> | ||
<p>Both selected sensitivity tuner constructs were made and submitted to the iGEM registry by the Cambridge 2009 team. | <p>Both selected sensitivity tuner constructs were made and submitted to the iGEM registry by the Cambridge 2009 team. | ||
</br></br> | </br></br> | ||
| Line 1,036: | Line 1,039: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | ||
| + | </br> | ||
<p>In order to implement these two sensitivity tuner variants into the Sensynova platform, designs were made by inserting the above parts between the two constructs forming the empty processor module of our framework.</p> | <p>In order to implement these two sensitivity tuner variants into the Sensynova platform, designs were made by inserting the above parts between the two constructs forming the empty processor module of our framework.</p> | ||
<img class="img-fluid border border-dark rounded" style="margin: 2%" src="https://static.igem.org/mediawiki/2017/9/95/T--Newcastle--Lais--ST--SBOL.png"></img> | <img class="img-fluid border border-dark rounded" style="margin: 2%" src="https://static.igem.org/mediawiki/2017/9/95/T--Newcastle--Lais--ST--SBOL.png"></img> | ||
| Line 1,058: | Line 1,062: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | ||
| + | </br> | ||
<p>The sensitivity tuners parts BBa_K274371 and BBa_K274381 were requested from the iGEM parts registry. Upon arrival, parts were <a href="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--ecoli_transformation_bb.pdf">transformed in DH5α <i>E. coli </i> cells</a>. Colonies were picked and cultures were prepared for <a href="https://static.igem.org/mediawiki/2017/e/e1/T--Newcastle--Miniprep.pdf">miniprepping</a>. Minipreps were <a href="https://static.igem.org/mediawiki/2017/1/13/T--Newcastle--digest.pdf">digested</a> with XbaI and PstI for BioBrick assembly [Protocol link]. | <p>The sensitivity tuners parts BBa_K274371 and BBa_K274381 were requested from the iGEM parts registry. Upon arrival, parts were <a href="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--ecoli_transformation_bb.pdf">transformed in DH5α <i>E. coli </i> cells</a>. Colonies were picked and cultures were prepared for <a href="https://static.igem.org/mediawiki/2017/e/e1/T--Newcastle--Miniprep.pdf">miniprepping</a>. Minipreps were <a href="https://static.igem.org/mediawiki/2017/1/13/T--Newcastle--digest.pdf">digested</a> with XbaI and PstI for BioBrick assembly [Protocol link]. | ||
</br></br> | </br></br> | ||
| Line 1,071: | Line 1,076: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
| + | </br> | ||
<p>Due to time constraints, we lacked the time to characterise these parts into the Sensynova platform within the lab. The parts <a href="http://parts.igem.org/Part:BBa_K2205024"> BBa_K2205024</a> and <a href="http://parts.igem.org/Part:BBa_K2205025"> BBa_K2205025</a>, the parts <a href="http://parts.igem.org/Part:BBa_K274371"> BBa_K274371 </a> and <a href="http://parts.igem.org/Part:BBa_K274381"> BBa_K274381 </a> respectively as processing units of the platform, were been submitted to the iGEM registry for future work and characterisation by future teams. | <p>Due to time constraints, we lacked the time to characterise these parts into the Sensynova platform within the lab. The parts <a href="http://parts.igem.org/Part:BBa_K2205024"> BBa_K2205024</a> and <a href="http://parts.igem.org/Part:BBa_K2205025"> BBa_K2205025</a>, the parts <a href="http://parts.igem.org/Part:BBa_K274371"> BBa_K274371 </a> and <a href="http://parts.igem.org/Part:BBa_K274381"> BBa_K274381 </a> respectively as processing units of the platform, were been submitted to the iGEM registry for future work and characterisation by future teams. | ||
</br></br> | </br></br> | ||
| Line 1,077: | Line 1,083: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
| + | </br> | ||
<p> iGEM Community. (2009). Team Cambridge 2009. [online] Available at: https://2009.igem.org/Team:Cambridge [Accessed 30 Oct. 2017]. | <p> iGEM Community. (2009). Team Cambridge 2009. [online] Available at: https://2009.igem.org/Team:Cambridge [Accessed 30 Oct. 2017]. | ||
</p> | </p> | ||
| Line 1,094: | Line 1,101: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| + | </br> | ||
<p>deGFP is a variant of Green Fluorescent Protein (GFP). It was initially designed by Shin and Noireaux (2010) for expression in cell-free protein synthesis (CFPS) systems and is more efficiently translated than other variants (e.g. eGFP). Through talks with other biosensor developers (for example, Chris French), and after reviewing legislation regarding the use of synthetic biology outside of the lab environment, the importance of CFPS systems as a chassis was highlighted. Despite its importance, CFPS systems can still suffer from some issues such as lower total protein synthesis than whole cells. By standardising and characterising a GFP variant which has been modified to have enhanced expression in these systems, it is hoped that CFPS will become a more attractive option for researchers. | <p>deGFP is a variant of Green Fluorescent Protein (GFP). It was initially designed by Shin and Noireaux (2010) for expression in cell-free protein synthesis (CFPS) systems and is more efficiently translated than other variants (e.g. eGFP). Through talks with other biosensor developers (for example, Chris French), and after reviewing legislation regarding the use of synthetic biology outside of the lab environment, the importance of CFPS systems as a chassis was highlighted. Despite its importance, CFPS systems can still suffer from some issues such as lower total protein synthesis than whole cells. By standardising and characterising a GFP variant which has been modified to have enhanced expression in these systems, it is hoped that CFPS will become a more attractive option for researchers. | ||
</br></br> | </br></br> | ||
| Line 1,099: | Line 1,107: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
| + | </br> | ||
<p>deGFP is a modified variant of eGFP developed by Shin and Noireaux which is more efficiently translated in CFPS systems. It was designed by truncating the N-terminal sequence and introducing silent mutations which removed internal ribosome binding like sequences. The C-terminal sequence is also truncated as this has been shown to not be necessary for maximal fluorescence (Li <i>et al</i>. 1997). By removing ribosome binding like sequences, Shin and Noireaux have reduced the amount of incorrect ribosome binding events and hence increased translation efficiency. The length of the protein also contributes to enhanced translation efficiency by reducing the time and resources required for this process to reach completion.</p> | <p>deGFP is a modified variant of eGFP developed by Shin and Noireaux which is more efficiently translated in CFPS systems. It was designed by truncating the N-terminal sequence and introducing silent mutations which removed internal ribosome binding like sequences. The C-terminal sequence is also truncated as this has been shown to not be necessary for maximal fluorescence (Li <i>et al</i>. 1997). By removing ribosome binding like sequences, Shin and Noireaux have reduced the amount of incorrect ribosome binding events and hence increased translation efficiency. The length of the protein also contributes to enhanced translation efficiency by reducing the time and resources required for this process to reach completion.</p> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | ||
| + | </br> | ||
<p>The deGFP sequence was taken from the Addgene database (Plasmid #40019). The sequence was found to have no illegal restriction sites (i.e. no EcoRI, XbaI, SpeI, or PstI sites). A strong, standard Anderson promoter (J23100) and RBS (B0034) was added before the deGFP sequence with biobrick scar sites between each part. A double terminator (B0015) was added after the deGFP sequence. The entire construct was flanked by 30 bp overhangs with the pSB1C3 plasmid, such that the construct could be Gibson assembled into a pSB1C3 plasmid digested with XbaI and SpeI (Figure 1). Extra bases were added between the overhangs and the construct so that once the part was assembled into the plasmid, the XbaI and SpeI sites could be regenerated and the biobrick prefix and suffix restored. This construct (J23100-deGFP) with the overhangs was submitted to IDT for synthesis as a gBlock.</p> | <p>The deGFP sequence was taken from the Addgene database (Plasmid #40019). The sequence was found to have no illegal restriction sites (i.e. no EcoRI, XbaI, SpeI, or PstI sites). A strong, standard Anderson promoter (J23100) and RBS (B0034) was added before the deGFP sequence with biobrick scar sites between each part. A double terminator (B0015) was added after the deGFP sequence. The entire construct was flanked by 30 bp overhangs with the pSB1C3 plasmid, such that the construct could be Gibson assembled into a pSB1C3 plasmid digested with XbaI and SpeI (Figure 1). Extra bases were added between the overhangs and the construct so that once the part was assembled into the plasmid, the XbaI and SpeI sites could be regenerated and the biobrick prefix and suffix restored. This construct (J23100-deGFP) with the overhangs was submitted to IDT for synthesis as a gBlock.</p> | ||
| Line 1,116: | Line 1,126: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | ||
| + | </br> | ||
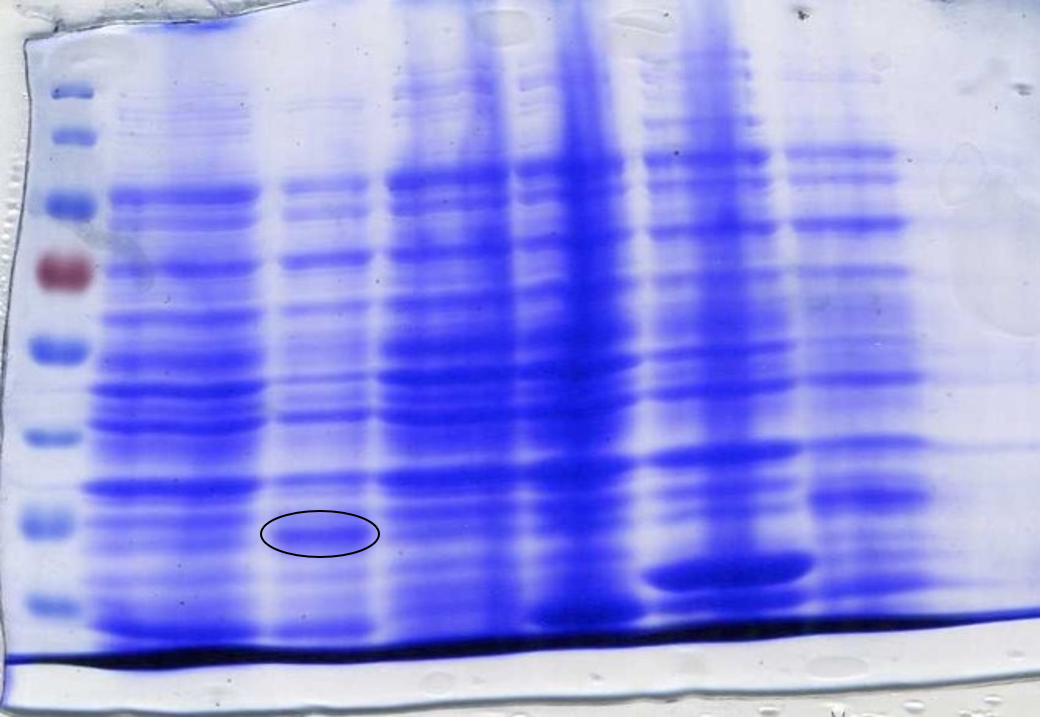
<p>The J23100-deGFP construct described above was Gibson assembled into a pSB1C3 plasmid using the NEB Hi-Fi assembly kit. To do this, pSB1C3 was <a href="https://static.igem.org/mediawiki/2017/1/13/T--Newcastle--digest.pdf">digested with XbaI and SpeI to create a linearised plasmid backbone</a>. The deGFP gBlock DNA was prepared according to the <a href="https://static.igem.org/mediawiki/2017/3/38/T--Newcastle--gBlock-HiFi.pdf">IDT protocol</a> and assembled into the linear plasmid backbone according to the <a href="https://static.igem.org/mediawiki/2017/3/38/T--Newcastle--gBlock-HiFi.pdf">NEB Hi-Fi Protocol</a>. The assembly mixture was then <a href="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--ecoli_transformation_bb.pdf">transformed</a> into commercial DH5α cells and incubated on chloramphenicol plates overnight. Colonies which were green under UV light were picked and grown in 5 mL LB broth <a href="https://static.igem.org/mediawiki/2017/7/73/T--Newcastle--cultures.pdf">overnight</a> before undergoing plasmid extraction [PROTOCOL]. Successful insertion of the J23100-deGFP construct into pSB1C3 was confirmed through a restriction digest with EcoRI and PstI followed by gel electrophoresis [PROTOCOL]. Figure 2 shows that the insert was successfully inserted as the double digest resulted in two linear bands at ~2100 bp (linear plasmid) and ~800 bp (deGFP). The DNA samples were then sent for sequencing to ensure that the construct was correct [DOWNLOAD LINK].</p> | <p>The J23100-deGFP construct described above was Gibson assembled into a pSB1C3 plasmid using the NEB Hi-Fi assembly kit. To do this, pSB1C3 was <a href="https://static.igem.org/mediawiki/2017/1/13/T--Newcastle--digest.pdf">digested with XbaI and SpeI to create a linearised plasmid backbone</a>. The deGFP gBlock DNA was prepared according to the <a href="https://static.igem.org/mediawiki/2017/3/38/T--Newcastle--gBlock-HiFi.pdf">IDT protocol</a> and assembled into the linear plasmid backbone according to the <a href="https://static.igem.org/mediawiki/2017/3/38/T--Newcastle--gBlock-HiFi.pdf">NEB Hi-Fi Protocol</a>. The assembly mixture was then <a href="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--ecoli_transformation_bb.pdf">transformed</a> into commercial DH5α cells and incubated on chloramphenicol plates overnight. Colonies which were green under UV light were picked and grown in 5 mL LB broth <a href="https://static.igem.org/mediawiki/2017/7/73/T--Newcastle--cultures.pdf">overnight</a> before undergoing plasmid extraction [PROTOCOL]. Successful insertion of the J23100-deGFP construct into pSB1C3 was confirmed through a restriction digest with EcoRI and PstI followed by gel electrophoresis [PROTOCOL]. Figure 2 shows that the insert was successfully inserted as the double digest resulted in two linear bands at ~2100 bp (linear plasmid) and ~800 bp (deGFP). The DNA samples were then sent for sequencing to ensure that the construct was correct [DOWNLOAD LINK].</p> | ||
| Line 1,130: | Line 1,141: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | ||
| + | </br> | ||
<p>The expression of deGFP was first tested in <i>E. coli</i> cells using an experimental procedure similar to that used in the Interlab study. Cells transformed with pSB1C3-J23100-deGFP were grown in 10 mL LB broth overnight and OD600 nm was measured. Culture was added to 3 separate falcon tubes and made up to 12 mL with LB with chloramphenicol such that the starting OD600 of the culture was approximately 0.02. This set-up was repeated with cells containing an identical plasmid and construct, except sfGFP was in place of deGFP. As a control, untransformed cells were also prepared identically except the LB did not contain chloramphenicol. Tubes with only LB and LB+chloramphenicol were also prepared as blanks. | <p>The expression of deGFP was first tested in <i>E. coli</i> cells using an experimental procedure similar to that used in the Interlab study. Cells transformed with pSB1C3-J23100-deGFP were grown in 10 mL LB broth overnight and OD600 nm was measured. Culture was added to 3 separate falcon tubes and made up to 12 mL with LB with chloramphenicol such that the starting OD600 of the culture was approximately 0.02. This set-up was repeated with cells containing an identical plasmid and construct, except sfGFP was in place of deGFP. As a control, untransformed cells were also prepared identically except the LB did not contain chloramphenicol. Tubes with only LB and LB+chloramphenicol were also prepared as blanks. | ||
</br></br> | </br></br> | ||
| Line 1,182: | Line 1,194: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
| + | </br> | ||
<p> | <p> | ||
We have successfully managed to BioBrick standardise deGFP for the first time, and have fully characterised this reporter, both in cells and in cell-free systems. As expected, this GFP variant showed fluorescence in both types of chassis. We also compared this GFP variant to superfolder GFP (sfGFP), which as far as we are aware has not been done before. We found that sfGFP had a higher fluoresence intensity in whole cells and CFPS systems compared to deGFP.<br /> | We have successfully managed to BioBrick standardise deGFP for the first time, and have fully characterised this reporter, both in cells and in cell-free systems. As expected, this GFP variant showed fluorescence in both types of chassis. We also compared this GFP variant to superfolder GFP (sfGFP), which as far as we are aware has not been done before. We found that sfGFP had a higher fluoresence intensity in whole cells and CFPS systems compared to deGFP.<br /> | ||
| Line 1,190: | Line 1,203: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
| + | </br> | ||
<p> | <p> | ||
Li X., Zhang G., Ngo N., Zhao X., Kain S.R., Huang C.C., (1997), Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence, <i>J. Biol. Chem.</i>, 272:28545–9, doi: 10.1074/jbc.272.45.28545<br /> | Li X., Zhang G., Ngo N., Zhao X., Kain S.R., Huang C.C., (1997), Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence, <i>J. Biol. Chem.</i>, 272:28545–9, doi: 10.1074/jbc.272.45.28545<br /> | ||
| Line 1,210: | Line 1,224: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
| + | </br> | ||
<p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. | <p>The Sensynova multicellular biosensor platform has been developed to overcome the <a href="https://2017.igem.org/Team:Newcastle/HP/Silver">limitations identified by our team</a> that hamper success in biosensor development. One of these limits regards the lack of modularity and reusability of the various components. Our platform design, based on the expression of three main modules (Detector, Processor and Reporter) by three <i>E.coli</i> strains in co-culture, allows the switch of possible variances for each module and the production of multiple customised biosensors. | ||
</br></br> | </br></br> | ||
| Line 1,215: | Line 1,230: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
| + | </br> | ||
<p>All three selected chromoproteins were made and submitted to the iGEM registry by the Uppsala 2013 team. | <p>All three selected chromoproteins were made and submitted to the iGEM registry by the Uppsala 2013 team. | ||
</br></br> | </br></br> | ||
| Line 1,235: | Line 1,251: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | ||
| + | </br> | ||
<p>In order to implement these three chromoprotein variants into the Sensynova platform, designs were made by replacing the sfGFP in the original reporter module with the parts detailed above that were ordered from the iGEM parts registry.</p> | <p>In order to implement these three chromoprotein variants into the Sensynova platform, designs were made by replacing the sfGFP in the original reporter module with the parts detailed above that were ordered from the iGEM parts registry.</p> | ||
<img src="https://static.igem.org/mediawiki/2017/8/8a/T--Newcastle--Lais--Chromo--SBOL.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/8/8a/T--Newcastle--Lais--Chromo--SBOL.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
| Line 1,260: | Line 1,277: | ||
</center></p></br> | </center></p></br> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Implementation </h2> | ||
| + | </br> | ||
<p>The chromoproteins aeBlue (BBa_K1033929), amajLime (BBa_K1033915) and spisPink (BBa_K1033925) parts were requested from the iGEM parts registry. Upon arrival, parts were <a href="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--ecoli_transformation_bb.pdf">transformed in DH5α <i>E. coli</i> cells</a>. Colonies were picked and overnight cultures were prepared for <a href="https://static.igem.org/mediawiki/2017/e/e1/T--Newcastle--Miniprep.pdf">miniprepping</a>. Minipreps were <a href="https://static.igem.org/mediawiki/2017/1/13/T--Newcastle--digest.pdf">digested</a> with XbaI and PstI for BioBrick assembly [Protocol link]. | <p>The chromoproteins aeBlue (BBa_K1033929), amajLime (BBa_K1033915) and spisPink (BBa_K1033925) parts were requested from the iGEM parts registry. Upon arrival, parts were <a href="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--ecoli_transformation_bb.pdf">transformed in DH5α <i>E. coli</i> cells</a>. Colonies were picked and overnight cultures were prepared for <a href="https://static.igem.org/mediawiki/2017/e/e1/T--Newcastle--Miniprep.pdf">miniprepping</a>. Minipreps were <a href="https://static.igem.org/mediawiki/2017/1/13/T--Newcastle--digest.pdf">digested</a> with XbaI and PstI for BioBrick assembly [Protocol link]. | ||
</br></br> | </br></br> | ||
| Line 1,267: | Line 1,285: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation </h2> | ||
| + | </br> | ||
<p> | <p> | ||
Initial testing of these three chromoprotein reporter variants was conducted by inoculating 1ml of LB containing the antibiotic Chloramphenicol with a colony from each colour proven by colony PCR and sequencing data to be correct and grown at 37° for 2 hours as well as a control of wildtype DH5α. | Initial testing of these three chromoprotein reporter variants was conducted by inoculating 1ml of LB containing the antibiotic Chloramphenicol with a colony from each colour proven by colony PCR and sequencing data to be correct and grown at 37° for 2 hours as well as a control of wildtype DH5α. | ||
| Line 1,326: | Line 1,345: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
| + | </br> | ||
<p> | <p> | ||
The qualitative results detailed above highlight the crucial advantage of our multicellular, modular framework, as it enables each component to be optimised avoiding any extra cloning steps. As each biosensor may be different and require specific designs and optimisation, easily choosing and changing modules and predicting <i>in silico</i> the bacterial community behaviour is essential for the development of new biosensors. | The qualitative results detailed above highlight the crucial advantage of our multicellular, modular framework, as it enables each component to be optimised avoiding any extra cloning steps. As each biosensor may be different and require specific designs and optimisation, easily choosing and changing modules and predicting <i>in silico</i> the bacterial community behaviour is essential for the development of new biosensors. | ||
| Line 1,334: | Line 1,354: | ||
</p> | </p> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
| + | </br> | ||
<p>Alieva, N., Konzen, K., Field, S., Meleshkevitch, E., Hunt, M., Beltran-Ramirez, V., Miller, D., Wiedenmann, J., Salih, A. and Matz, M. (2008). Diversity and Evolution of Coral Fluorescent Proteins. PLoS ONE, 3(7), p.e2680. | <p>Alieva, N., Konzen, K., Field, S., Meleshkevitch, E., Hunt, M., Beltran-Ramirez, V., Miller, D., Wiedenmann, J., Salih, A. and Matz, M. (2008). Diversity and Evolution of Coral Fluorescent Proteins. PLoS ONE, 3(7), p.e2680. | ||
</br></br> | </br></br> | ||
Revision as of 17:13, 1 November 2017
spacefill
spacefill
Our Experimental Results
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project






























































 The
The