| Line 119: | Line 119: | ||
<h1 style="font-weight:normal; font-family: Rubik; margin: 0">Our Experimental Results</h1> | <h1 style="font-weight:normal; font-family: Rubik; margin: 0">Our Experimental Results</h1> | ||
| − | < | + | |
| + | |||
| + | <div class="jumbotron rounded" style="background-color: #fddd85; border: 4px solid #171717; margin-top: 2%; margin-bottom: 2%"> | ||
| + | <h1 class="display-3" style="color: #171717">Key Achievements</h1> | ||
| + | <p class="lead" style="color: #171717">A condensed list of our most notable results</p> | ||
| + | <hr style="color: #171717"> | ||
| + | <ul style="list-style: none; color: #171717"> | ||
| + | <li style="font-family: Rubik">- Designed a novel framework for biosensor development</li> | ||
| + | <li style="font-family: Rubik">- Proved that multicellular biosensors are able to co-ordinate responses to input molecules through a proof-of-concept IPTG responsive biosensor</li> | ||
| + | <li style="font-family: Rubik">- Successful characterisation of a transpose-based “stand-by switch” capable of producing eforRed in the “OFF” state, and C4 AHL in the “ON” state</li> | ||
| + | <li style="font-family: Rubik">- Used a Design of Experiments approach to successfully optimise a cell-free system</li> | ||
| + | <li style="font-family: Rubik">- Improved the BLANK plasmid for promoter screening</li> | ||
| + | <li style="font-family: Rubik">- Expressed and characterised Sarcosine Oxidase, showing successful degradation of sarcosine to formaldehyde</li> | ||
| + | <li style="font-family: Rubik">- Designed, and began to construct, a variety of framework compatible systems, including a synthetic promoter library</li> | ||
| + | <li style="font-family: Rubik">- Determined optimal cell ratios from our <a href="https://2017.igem.org/Team:Newcastle/Model#sim">multicellular model</a></li> | ||
| + | <li style | ||
| + | </ul> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
<p> | <p> | ||
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.<br /> | Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.<br /> | ||
| Line 206: | Line 225: | ||
<center> | <center> | ||
<p>Multicellular Framework Testing</p> | <p>Multicellular Framework Testing</p> | ||
| − | |||
<a href="https://2017.igem.org/Team:Newcastle/Results#myTab"><img onClick="showTab('framework')" title="Full Framework in Cells" id="framework_res" class="hoverable hover_large" src="https://static.igem.org/mediawiki/2017/7/7b/T--Newcastle--BB_flask_results.png"/></a> | <a href="https://2017.igem.org/Team:Newcastle/Results#myTab"><img onClick="showTab('framework')" title="Full Framework in Cells" id="framework_res" class="hoverable hover_large" src="https://static.igem.org/mediawiki/2017/7/7b/T--Newcastle--BB_flask_results.png"/></a> | ||
</center> | </center> | ||
| Line 290: | Line 308: | ||
<center> | <center> | ||
<p>Framework in Cell Free Protein Synthesis Systems</p> | <p>Framework in Cell Free Protein Synthesis Systems</p> | ||
| − | |||
<a href="https://2017.igem.org/Team:Newcastle/Results#myTab"><img onClick="showTab('cellfree')" id="cellfree_res" title="Cell Free Protein Synthesis Systems" class="hoverable hover_large" src="https://static.igem.org/mediawiki/2017/b/b6/T--Newcastle--BB_CFPS_results.png"/></a> | <a href="https://2017.igem.org/Team:Newcastle/Results#myTab"><img onClick="showTab('cellfree')" id="cellfree_res" title="Cell Free Protein Synthesis Systems" class="hoverable hover_large" src="https://static.igem.org/mediawiki/2017/b/b6/T--Newcastle--BB_CFPS_results.png"/></a> | ||
</center> | </center> | ||
| Line 469: | Line 486: | ||
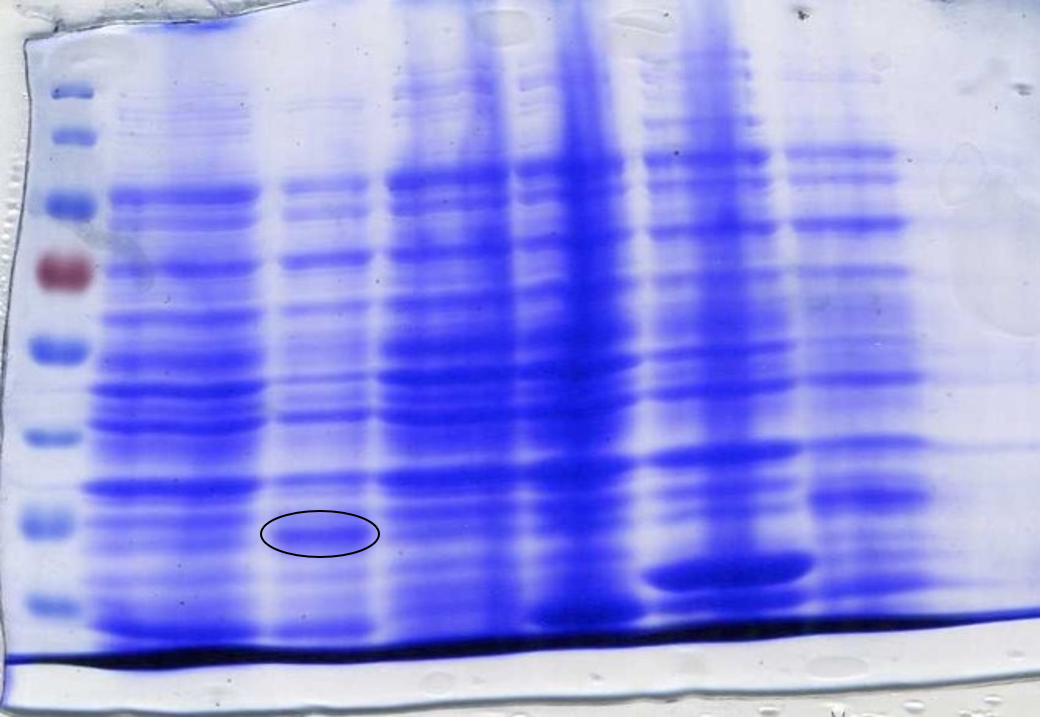
<p>To determine whether the now correct SOX had been successfully expressed another SDS-Page gel was performed. After inducing, harvesting and washing the cells 1 ml was taken from each culture to be loaded into the gel. The cells were lysed using lysozyme and boiled for 3 minutes at 100°C loading 10 µl into the gel (Figure 7). | <p>To determine whether the now correct SOX had been successfully expressed another SDS-Page gel was performed. After inducing, harvesting and washing the cells 1 ml was taken from each culture to be loaded into the gel. The cells were lysed using lysozyme and boiled for 3 minutes at 100°C loading 10 µl into the gel (Figure 7). | ||
</br></br> | </br></br> | ||
| − | <div class="SOX"><img src="https://static.igem.org/mediawiki/2017/8/89/T--Newcastle--Correct_sox_protein_gel_2.png" width="30%" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt="" class="img-fluid border border-dark rounded mx-auto d-block"/> | + | <div class="SOX"><img src="https://static.igem.org/mediawiki/2017/8/89/T--Newcastle--Correct_sox_protein_gel_2.png" width="30%" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt="" class="img-fluid border border-dark rounded mx-auto d-block"/> |
<br /> | <br /> | ||
<p class="legend"><center><strong>Figure 7:</strong> Sarcosine Oxidase expression was induced by adding 40 µl of 100 mM IPTG. Lane 1: 6 µl ladder, Lane 2: 10 µl sfGFP, Lane 3: BL21-DE3, Lane 4: 10µl SOX 1, Lane 5: 10 µl SOX 2, Lane 6: 10 µl SOX 3, Lane 7: 10 µl SOX 4, Lane 8: 10 µl SOX 5, Lane 9: 10 µl SOX 6, Lane 10: 6 µl ladder. Circled bands show sarcosine oxidase at ~42 kDa, the expected molecular weight.</p></center> | <p class="legend"><center><strong>Figure 7:</strong> Sarcosine Oxidase expression was induced by adding 40 µl of 100 mM IPTG. Lane 1: 6 µl ladder, Lane 2: 10 µl sfGFP, Lane 3: BL21-DE3, Lane 4: 10µl SOX 1, Lane 5: 10 µl SOX 2, Lane 6: 10 µl SOX 3, Lane 7: 10 µl SOX 4, Lane 8: 10 µl SOX 5, Lane 9: 10 µl SOX 6, Lane 10: 6 µl ladder. Circled bands show sarcosine oxidase at ~42 kDa, the expected molecular weight.</p></center> | ||
</div> | </div> | ||
</br></br> | </br></br> | ||
| − | + | <p>To test for the presence of formaldehyde, and to demonstrate this part works, larger cultures were grown following the aforementioned protocols, and the cells harvested, washed and lysed by sonication. 0 µl, 20 µl, 200 µl and 2 ml of Glyphosate at 10 mg/L concentration was added to the cell lysate and incubated at 37°C. Every 2.5 hours the lysate was tested for the presence of formaldehyde with commercial <a href="http://www.sigmaaldrich.com/catalog/product/sial/37072?lang=en®ion=GB">formaldehyde testing strips</a>.</p> | |
| + | </br></br> | ||
| + | <p>After 8 hours of testing and left overnight, none of the samples had produced formaldehyde according to the testing strips. The testing strips detect a minimum formaldehyde concentration of 10 mg/L, so it was possible that formaldehyde had been produced but that there was too little of it to detect with the strips.</p> | ||
| + | </br></br> | ||
| + | <p> We decided to add Sarcosine instead of Glyphosate to determine whether the part was working. Everything was repeated the same but instead we added 0 µl, 50 µl and 200 µl of Sarcosine at 0.9 g/50 ml (Figure 8). | ||
</br></br> | </br></br> | ||
<div class="SOX"><img src="https://static.igem.org/mediawiki/2017/4/4b/T--Newcastle--SOX_testing.JPG" width="30%" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt="" class="img-fluid border border-dark rounded mx-auto d-block"/> | <div class="SOX"><img src="https://static.igem.org/mediawiki/2017/4/4b/T--Newcastle--SOX_testing.JPG" width="30%" style="background-color:white; margin-right: 2%; margin-bottom: 2%;" alt="" class="img-fluid border border-dark rounded mx-auto d-block"/> | ||
| Line 481: | Line 502: | ||
</div> | </div> | ||
</br></br> | </br></br> | ||
| − | <p>This shows SOX works as expected, however there is | + | <p>This shows SOX works as expected, however there is leaky expression as formaldehyde is produced when no IPTG is added.</p> |
</p> | </p> | ||
</br></br> | </br></br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
</br> | </br> | ||
| − | <p><i>E. coli</i> cells naturally have the C-P lyase pathway which degrades glyphosate into sarcosine. The fact that formaldehyde was produced when | + | <p><i>E. coli</i> cells naturally have the C-P lyase pathway which degrades glyphosate into sarcosine. The fact that no formaldehyde was produced when glyphosate was added, but was when sarcosine was added, indicates that we have not overexpressed the C-P lyase pathway enough to produce enough sarcosine for SOX to convert into formaldehyde to be detected. |
</br></br> | </br></br> | ||
<p>Due to time constraints, we were unable to produce an <i>in vivo</i> formaldehyde detector variant of the Sensynova framework. Future characterisation of this part would include using the platform customised as a formaldehyde biosensor in order to sense compound produce and therefore creating a biosensor of glyphosate. | <p>Due to time constraints, we were unable to produce an <i>in vivo</i> formaldehyde detector variant of the Sensynova framework. Future characterisation of this part would include using the platform customised as a formaldehyde biosensor in order to sense compound produce and therefore creating a biosensor of glyphosate. | ||
| Line 538: | Line 555: | ||
<center><b>Figure 2:</b> Graph Indicating the Most Frequent Spacer Between -35 and -10 Regions Found in <i>E. coli</i> Promoters. This image was taken from Harley and Reynolds (1987).</center> | <center><b>Figure 2:</b> Graph Indicating the Most Frequent Spacer Between -35 and -10 Regions Found in <i>E. coli</i> Promoters. This image was taken from Harley and Reynolds (1987).</center> | ||
</p> | </p> | ||
| − | </br> | + | </br> |
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | ||
</br> | </br> | ||
| Line 584: | Line 601: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
</br> | </br> | ||
| − | <p>Though we have generated a sizable library of promoters of varying strengths and functions, we lacked the time to complete its characterization by the screening against targeted molecules. | + | <p>Though we have generated a sizable library of promoters of varying strengths and functions, we lacked the time to complete its characterization by the screening against targeted molecules. |
<br/><br/> | <br/><br/> | ||
Due to time constraints, we also lacked the time to characterise these parts into the Sensynova platform within the lab. | Due to time constraints, we also lacked the time to characterise these parts into the Sensynova platform within the lab. | ||
| Line 627: | Line 644: | ||
<img src="https://static.igem.org/mediawiki/2017/b/b1/Vava1aa.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/b/b1/Vava1aa.png" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
| − | <center><b>Figure 1:</b> | + | <center><b>Figure 1:</b> <a href="http://parts.igem.org/Part:BBa_J33201">BBa_J33201</a> design.</center></p> |
</br> | </br> | ||
| Line 640: | Line 657: | ||
<td> | <td> | ||
<img src="https://static.igem.org/mediawiki/2017/d/db/Vave2.jpg" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | <img src="https://static.igem.org/mediawiki/2017/d/db/Vave2.jpg" class="img-fluid border border-dark rounded" style="margin: 2%; max-width: 70%"> | ||
| − | <div><p class="legend"><center><strong><b>Figure 2:</b> </strong> | + | <div><p class="legend"><center><strong><b>Figure 2:</b> </strong> The new part <a href="http://parts.igem.org/Part:BBa_K2205022">BBa_K2205022</a> based on the previous Arsenic detector design <a href="http://parts.igem.org/Part:BBa_J33201">BBa_J33201</a>, implemented by the Sensynova part coding for the connector 1, <a href="http://parts.igem.org/Part:BBa_K2205008">BBa_K2205008</a>.</p></center></div> |
</td> | </td> | ||
| Line 790: | Line 807: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation</h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Characterisation</h2> | ||
</br> | </br> | ||
| − | <p> A preliminary qualitative assay was carried out as an initial test for this construct. Co-cultures of Psicose detector, processor unit and sfGFP reporter (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2205015">BBa_K2205015</a>) were inoculated and grown overnight in LB+chloramphenicol (12.5 ng/ul). | + | <p> A preliminary qualitative assay was carried out as an initial test for this construct. Co-cultures of Psicose detector, processor unit and sfGFP reporter (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2205015">BBa_K2205015</a>) were inoculated and grown overnight in LB+chloramphenicol (12.5 ng/ul). |
</br></br> | </br></br> | ||
The day after the cultures were diluted at OD600 0.1 and mixed together to obtain co-cultures with ratio 1:1:13 (detector:processor:reporter). The samples were supplemented with 33.22 mM Psicose to induce the expression of quorum sensing molecules and eventually achieve the reporter visualisation (Figures 8). </p> | The day after the cultures were diluted at OD600 0.1 and mixed together to obtain co-cultures with ratio 1:1:13 (detector:processor:reporter). The samples were supplemented with 33.22 mM Psicose to induce the expression of quorum sensing molecules and eventually achieve the reporter visualisation (Figures 8). </p> | ||
| Line 814: | Line 831: | ||
<hr> | <hr> | ||
| − | + | ||
<h1 style="font-family: Rubik"> Formaldehyde <button class="btn btn-primary collapsed" type="button" data-toggle="collapse" data-target="#formaldehyde" aria-expanded="false" aria-controls="formaldehyde" style="margin-left: 1%"></button></h1> | <h1 style="font-family: Rubik"> Formaldehyde <button class="btn btn-primary collapsed" type="button" data-toggle="collapse" data-target="#formaldehyde" aria-expanded="false" aria-controls="formaldehyde" style="margin-left: 1%"></button></h1> | ||
<div id="formaldehyde" class="collapse"> | <div id="formaldehyde" class="collapse"> | ||
| Line 820: | Line 837: | ||
<h2 style="font-size: 1em"> BioBricks used: <a href="http://parts.igem.org/Part:BBa_K2205029">BBa_K2205029 (New)</a>, <a href="http://parts.igem.org/Part:BBa_K2205030">BBa_K2205030 (New)</a>, <a href="http://parts.igem.org/Part:BBa_K749021">BBa_K749021(TMU-Tokyo 2012 )</a> </h2> | <h2 style="font-size: 1em"> BioBricks used: <a href="http://parts.igem.org/Part:BBa_K2205029">BBa_K2205029 (New)</a>, <a href="http://parts.igem.org/Part:BBa_K2205030">BBa_K2205030 (New)</a>, <a href="http://parts.igem.org/Part:BBa_K749021">BBa_K749021(TMU-Tokyo 2012 )</a> </h2> | ||
</br> | </br> | ||
| − | + | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
</br> | </br> | ||
| Line 831: | Line 848: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Background Information </h2> | ||
</br> | </br> | ||
| − | <p>The formaldehyde biosensor, part BBa_K749021, was selected was originally made and submitted to the iGEM registry by the TMU-Tokyo 2012 team. | + | <p>The formaldehyde biosensor, part BBa_K749021, was selected was originally made and submitted to the iGEM registry by the TMU-Tokyo 2012 team. |
</br></br> | </br></br> | ||
| − | This part was chosen as a variant to the detector module present in the Sensynova platform due to the fact that our adaptor module present in the framework, Sarcosine Oxidase, was made in order to convert glyphosate into formaldehyde, in order to overcome the limitation in the detection of glyphosate due to its little-known knowledge. | + | This part was chosen as a variant to the detector module present in the Sensynova platform due to the fact that our adaptor module present in the framework, Sarcosine Oxidase, was made in order to convert glyphosate into formaldehyde, in order to overcome the limitation in the detection of glyphosate due to its little-known knowledge. |
</br> | </br> | ||
<center><img src="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--Lais--FO--Ruler.png" class="img-fluid border border-dark rounded" style="margin: 2%"></center> | <center><img src="https://static.igem.org/mediawiki/2017/1/1f/T--Newcastle--Lais--FO--Ruler.png" class="img-fluid border border-dark rounded" style="margin: 2%"></center> | ||
| Line 852: | Line 869: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Design Stage </h2> | ||
</br> | </br> | ||
| − | <p>In order to implement the Formaldehyde biosensor variant to the Sensynova platform, a design was created by replacing the IPTG sensing system in the original detector module with the construct detailed above, creating part <a href="http://parts.igem.org/Part:BBa_K2205030">BBa_K2205030 </a>. | + | <p>In order to implement the Formaldehyde biosensor variant to the Sensynova platform, a design was created by replacing the IPTG sensing system in the original detector module with the construct detailed above, creating part <a href="http://parts.igem.org/Part:BBa_K2205030">BBa_K2205030 </a>. |
</br></br> | </br></br> | ||
We chose to redesign the Formaldehyde biosensor detailed above to mirror the design used when producing the Psicose detector variant. The system detailed in the image below is made up of the constitutive promoter present within the platform triggering transcription of the FrmR repressing the PfrmR and subsequently the connector 1 of the Sensynova platform. We have also replaced the colour output present in the TMU-Tokyo design, we have added our part <a href="http://parts.igem.org/Part:BBa_K2205008">BBa_K2205008</a>, which produces our first connector in order to trigger a response from following modules of the Sensynova platform in the presence of Formaldehyde.</p> | We chose to redesign the Formaldehyde biosensor detailed above to mirror the design used when producing the Psicose detector variant. The system detailed in the image below is made up of the constitutive promoter present within the platform triggering transcription of the FrmR repressing the PfrmR and subsequently the connector 1 of the Sensynova platform. We have also replaced the colour output present in the TMU-Tokyo design, we have added our part <a href="http://parts.igem.org/Part:BBa_K2205008">BBa_K2205008</a>, which produces our first connector in order to trigger a response from following modules of the Sensynova platform in the presence of Formaldehyde.</p> | ||
| Line 882: | Line 899: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%">Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%">Conclusions and Future Work </h2> | ||
</br> | </br> | ||
| − | <p>Due to time constraints, we lacked the time to synthesise, implement and characterise this part into the Sensynova platform within the lab. Future work on this part would include characterisation <i>in vivo</i> guided by the modelling of the framework when customised as a formaldehyde biosensor and testing against the Sarcosine Oxidase adaptor module currently present in the framework. | + | <p>Due to time constraints, we lacked the time to synthesise, implement and characterise this part into the Sensynova platform within the lab. Future work on this part would include characterisation <i>in vivo</i> guided by the modelling of the framework when customised as a formaldehyde biosensor and testing against the Sarcosine Oxidase adaptor module currently present in the framework. |
</p> | </p> | ||
</br> | </br> | ||
| Line 893: | Line 910: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | ||
<div class="tab-pane fade" id="nav-processor" role="tabpanel" aria-labelledby="nav-processor-tab"> | <div class="tab-pane fade" id="nav-processor" role="tabpanel" aria-labelledby="nav-processor-tab"> | ||
</br> | </br> | ||
| Line 1,877: | Line 1,894: | ||
</div> | </div> | ||
</br> | </br> | ||
| + | |||
| + | |||
| + | |||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
| + | </br> | ||
<p>This study has begun multifactorial analysis on the components of the supplemental solution for cell free protein synthesis systems. It has provided evidence that some supplements have a greater effect on a systems protein synthesis activity than others, and that the important factors may differ between cell extract batches. The ability to use a Design of Experiments approach towards the optimisation of CFPS systems has also been demonstrated. While this study has provided evidence towards these claims, further work should be performed to validate the findings. A DoE screening design for the supplements of CFPS systems should be used on the same cell extract batch repeatedly. This will help confirm that the screening models derived from the experimental design data are accurate. The screening design should also be performed on many different batches of at least moderately active cell extracts to confirm that important supplements do differ between batches. | <p>This study has begun multifactorial analysis on the components of the supplemental solution for cell free protein synthesis systems. It has provided evidence that some supplements have a greater effect on a systems protein synthesis activity than others, and that the important factors may differ between cell extract batches. The ability to use a Design of Experiments approach towards the optimisation of CFPS systems has also been demonstrated. While this study has provided evidence towards these claims, further work should be performed to validate the findings. A DoE screening design for the supplements of CFPS systems should be used on the same cell extract batch repeatedly. This will help confirm that the screening models derived from the experimental design data are accurate. The screening design should also be performed on many different batches of at least moderately active cell extracts to confirm that important supplements do differ between batches. | ||
</br></br> | </br></br> | ||
| Line 1,884: | Line 1,905: | ||
</br> | </br> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
| + | </br> | ||
<p> | <p> | ||
Algranati, I. D. & Goldemberg, S. H., 1977. Polyamines and their role in protein synthesis. <i>Trends in Biochem. Sci.</i>, 2(12), pp. 272-274.<br /> | Algranati, I. D. & Goldemberg, S. H., 1977. Polyamines and their role in protein synthesis. <i>Trends in Biochem. Sci.</i>, 2(12), pp. 272-274.<br /> | ||
Revision as of 19:55, 1 November 2017
spacefill
spacefill
Our Experimental Results
Key Achievements
A condensed list of our most notable results
- - Designed a novel framework for biosensor development
- - Proved that multicellular biosensors are able to co-ordinate responses to input molecules through a proof-of-concept IPTG responsive biosensor
- - Successful characterisation of a transpose-based “stand-by switch” capable of producing eforRed in the “OFF” state, and C4 AHL in the “ON” state
- - Used a Design of Experiments approach to successfully optimise a cell-free system
- - Improved the BLANK plasmid for promoter screening
- - Expressed and characterised Sarcosine Oxidase, showing successful degradation of sarcosine to formaldehyde
- - Designed, and began to construct, a variety of framework compatible systems, including a synthetic promoter library
- - Determined optimal cell ratios from our multicellular model
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project
 |
 |
 |
 |
 |
 |
 |
 |






























































 The
The