| (17 intermediate revisions by 5 users not shown) | |||
| Line 110: | Line 110: | ||
.result_picture td p { | .result_picture td p { | ||
text-align: center; | text-align: center; | ||
| + | } | ||
| + | |||
| + | .keyres_hide { | ||
| + | display: none; | ||
| + | } | ||
| + | |||
| + | #key_res:hover { | ||
| + | cursor:pointer; | ||
} | } | ||
| Line 119: | Line 127: | ||
<h1 class="display 4" style="font-family: Rubik; margin: 0">Our Experimental Results</h1> | <h1 class="display 4" style="font-family: Rubik; margin: 0">Our Experimental Results</h1> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <div class="jumbotron rounded" style="background-color: #e8e8e8; border: 1px solid #222222; padding: 10px !important;"> | ||
| + | <h2 id="key_res" class="display-5 keyres_show" style="color: #222222">Key Achievements - click to show</h2> | ||
| + | <hr class="keyres_hide" style="color: #222222"> | ||
| + | <ul class="keyres_hide" style="color: #222222"> | ||
| + | |||
| + | |||
| + | <li class="keyres_hide" style="font-family: Rubik">Demonstrated that biosensors can be successfully split into three modules</li> | ||
| + | |||
| + | <li class="keyres_hide" style="font-family: Rubik">Produced biosensor variants by co-culturing different module variants together</li> | ||
| + | |||
| + | <li class="keyres_hide" style="font-family: Rubik">Used 3D spatial modelling to begin optimisation of a multicellular biosensor</li> | ||
| + | |||
| + | <li class="keyres_hide" style="font-family: Rubik">Characterised a 'standby switch' based on an improved part <a href="http://parts.igem.org/Part:BBa_K1632007">(BBa_K1632007)</a></li> | ||
| + | |||
| + | <li class="keyres_hide" style="font-family: Rubik">Demonstrated that a Design of Experiments approach can be used to optimise cell-free systems</li> | ||
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</ul> | </ul> | ||
</div> | </div> | ||
| Line 402: | Line 415: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Rationale and Aim </h2> | ||
</br> | </br> | ||
| − | <p>Glyphosate is a herbicide that works by blocking the activity of the enzyme enolpyruvylshikimate-3-phosphate synthase (EPSPS), which converts carbohydrates derived from glycolysis and the pentose phosphate pathway to plant metabolites and aromatic amino acids. | + | <p>Glyphosate is a herbicide that works by blocking the activity of the enzyme enolpyruvylshikimate-3-phosphate synthase (EPSPS), which converts carbohydrates derived from glycolysis and the pentose phosphate pathway to plant metabolites and aromatic amino acids. It was identified as a biosensor target through our <a href="https://2017.igem.org/Team:Newcastle/HP/Gold_Integrated">human practices</a> when we attended the N8 conference. |
</br></br> | </br></br> | ||
We attempted to design a system capable of glyphosate detection. With little information regarding mechanisms of glyphosate interactions within the cell, we could not identify a simple system in which a responsive transcription factor was able to affect the production of a reporter gene. This is a common issue in many biosensor projects. | We attempted to design a system capable of glyphosate detection. With little information regarding mechanisms of glyphosate interactions within the cell, we could not identify a simple system in which a responsive transcription factor was able to affect the production of a reporter gene. This is a common issue in many biosensor projects. | ||
| − | To show the adaptor in action we chose to develop a part that would measure the level of glyphosate through the production of formaldehyde. There are known sensors for formaldehyde such as | + | To show the adaptor in action we chose to develop a part that would measure the level of glyphosate through the production of formaldehyde. There are known sensors for formaldehyde such as <a href="https://2012.igem.org/Team:TMU-Tokyo">Tokyo’s 2012 biosensor</a>. Our design relies on the natural biochemical systems, the c-p lyase pathways, in <i>E. coli</i> to convert glyphosate to sarcosine. We then designed a part, SOX, based on the production of the enzyme sarcosine oxidase, encoded by <i>soxA</i> to convert sarcosine to formaldehyde ready for detection by a formaldehyde producing input module. |
</br></br> | </br></br> | ||
| Line 500: | Line 513: | ||
<p>Formaldehyde was detected, showing that SOX works as expected, however there is slight leaky expression as formaldehyde is produced when no IPTG is added.</p> | <p>Formaldehyde was detected, showing that SOX works as expected, however there is slight leaky expression as formaldehyde is produced when no IPTG is added.</p> | ||
</p> | </p> | ||
| − | + | </br> | |
| − | <p> We also decided to add Glyphosate to determine the efficiency of the C-P Lyase pathway. Everything was repeated the same but instead we added 0 µl, 20 µl, 200 µl and 2 ml of glyphosate at 10mg/L. | + | <p> We also decided to add Glyphosate to determine the efficiency of the native C-P Lyase pathway. Everything was repeated the same but instead we added 0 µl, 20 µl, 200 µl and 2 ml of glyphosate at 10mg/L. After 8 hours of testing and left overnight, none of the samples had produced formaldehyde according to the testing strips. The testing strips detect a minimum formaldehyde concentration of 10 mg/L, so it was possible that formaldehyde had been produced but that there was too little of it to detect with the strips.</p> |
| − | + | ||
| − | + | ||
| − | + | </br> | |
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> Conclusions and Future Work </h2> | ||
| Line 609: | Line 620: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
</br> | </br> | ||
| − | <p>Becker, N., Peters, J., Lionberger, T. and Maher, L. (2012). Mechanism of promoter repression by Lac repressor–DNA loops. Nucleic Acids Research, 41(1), pp.156-166. <br/> | + | <p>Becker, N., Peters, J., Lionberger, T. and Maher, L. (2012). Mechanism of promoter repression by Lac repressor–DNA loops. Nucleic Acids Research, 41(1), pp.156-166. <br/><br/> |
DeBoer, H. (1985). Microbial hybrid promoters. US4551433 A. | DeBoer, H. (1985). Microbial hybrid promoters. US4551433 A. | ||
<br/><br/> | <br/><br/> | ||
| Line 733: | Line 744: | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
</br> | </br> | ||
| − | <p>Brenner K, Karing D, Weiss R, Arnold F (2007) Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA 104(44): 17300 - 17304 </br> de Mora K, Joshi N, Balint BL, Ward FB, Elfick A, French CE (2011) A pH-based biosensor for detection of arsenic in drinking water. Anal Bioanal Chem 400(4):1031-9 (Epub 2011 Mar 27).</p> | + | <p>Brenner K, Karing D, Weiss R, Arnold F (2007) Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA 104(44): 17300 - 17304 <br/><br/> de Mora K, Joshi N, Balint BL, Ward FB, Elfick A, French CE (2011) A pH-based biosensor for detection of arsenic in drinking water. Anal Bioanal Chem 400(4):1031-9 (Epub 2011 Mar 27).</p> |
</div> | </div> | ||
| Line 1,249: | Line 1,260: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | Li X., Zhang G., Ngo N., Zhao X., Kain S.R., Huang C.C., (1997), Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence, <i>J. Biol. Chem.</i>, 272:28545–9, doi: 10.1074/jbc.272.45.28545<br /> | + | Li X., Zhang G., Ngo N., Zhao X., Kain S.R., Huang C.C., (1997), Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence, <i>J. Biol. Chem.</i>, 272:28545–9, doi: 10.1074/jbc.272.45.28545 <br/><br/> |
Shin, J., and Noireaux, V., (2010), Efficient Cell-Free Expression with the Endogenous <i>E. coli</i> RNA Polymerase and Sigma Factor 70, <i>J. Biol. Eng.</i>, 4:8, doi: 10.1186/1754-1611-4-8<br /> | Shin, J., and Noireaux, V., (2010), Efficient Cell-Free Expression with the Endogenous <i>E. coli</i> RNA Polymerase and Sigma Factor 70, <i>J. Biol. Eng.</i>, 4:8, doi: 10.1186/1754-1611-4-8<br /> | ||
| Line 1,903: | Line 1,914: | ||
<p>This study has begun multifactorial analysis on the components of the supplemental solution for cell free protein synthesis systems. It has provided evidence that some supplements have a greater effect on a systems protein synthesis activity than others, and that the important factors may differ between cell extract batches. The ability to use a Design of Experiments approach towards the optimisation of CFPS systems has also been demonstrated. While this study has provided evidence towards these claims, further work should be performed to validate the findings. A DoE screening design for the supplements of CFPS systems should be used on the same cell extract batch repeatedly. This will help confirm that the screening models derived from the experimental design data are accurate. The screening design should also be performed on many different batches of at least moderately active cell extracts to confirm that important supplements do differ between batches. | <p>This study has begun multifactorial analysis on the components of the supplemental solution for cell free protein synthesis systems. It has provided evidence that some supplements have a greater effect on a systems protein synthesis activity than others, and that the important factors may differ between cell extract batches. The ability to use a Design of Experiments approach towards the optimisation of CFPS systems has also been demonstrated. While this study has provided evidence towards these claims, further work should be performed to validate the findings. A DoE screening design for the supplements of CFPS systems should be used on the same cell extract batch repeatedly. This will help confirm that the screening models derived from the experimental design data are accurate. The screening design should also be performed on many different batches of at least moderately active cell extracts to confirm that important supplements do differ between batches. | ||
</br></br> | </br></br> | ||
| − | Following the above, a surface response design could be used for all commonly important supplements of the CFPS system to determine its effectiveness at optimising CFPS activity. The information could also be used to determine commonly unimportant supplements so they can be eliminated from the supplement solution, hence decreasing the cost per reaction.</p> | + | Following the above, a surface response design could be used for all commonly important supplements of the CFPS system to determine its effectiveness at optimising CFPS activity. The information could also be used to determine commonly unimportant supplements so they can be eliminated from the supplement solution, hence decreasing the cost per reaction. Furthermore, the full Sensynova system can be expressed in cell-free.</p> |
</br> | </br> | ||
<h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | <h2 style="font-family: Rubik; text-align: left; margin-top: 1%"> References </h2> | ||
</br> | </br> | ||
<p> | <p> | ||
| − | Algranati, I. D. & Goldemberg, S. H., 1977. Polyamines and their role in protein synthesis. <i>Trends in Biochem. Sci.</i>, 2(12), pp. 272-274.<br /> | + | Algranati, I. D. & Goldemberg, S. H., 1977. Polyamines and their role in protein synthesis. <i>Trends in Biochem. Sci.</i>, 2(12), pp. 272-274. <br/><br/> |
| − | Anderson, M. J. & Whitcomb, P. J., 2010. Design of Experiments. In: Kirk-Othmer Encyclopedia of Chemical Technology. <i>s.l.:John Wiley & Sons, Inc</i>, pp. 1-22. <br /> | + | Anderson, M. J. & Whitcomb, P. J., 2010. Design of Experiments. In: Kirk-Othmer Encyclopedia of Chemical Technology. <i>s.l.:John Wiley & Sons, Inc</i>, pp. 1-22. <br/><br/> |
| − | Borg, A. & Ehrenberg, M., 2015. Determinants of the Rate of mRNA Translocation in Bacterial Protein Synthesis. <i>J. Mol. Biol.</i>, 427(9), pp. 1835-1847.<br /> | + | Borg, A. & Ehrenberg, M., 2015. Determinants of the Rate of mRNA Translocation in Bacterial Protein Synthesis. <i>J. Mol. Biol.</i>, 427(9), pp. 1835-1847. <br/><br/> |
| − | Carlson, E. D., Gan, R., Hodgman, C. E. & Jewett, M. C., 2012. Cell-Free Protein Synthesis: Applications Come of Age. <i>Biotechnol. Adv.</i>, 30(5), pp. 1185-1194.<br /> | + | Carlson, E. D., Gan, R., Hodgman, C. E. & Jewett, M. C., 2012. Cell-Free Protein Synthesis: Applications Come of Age. <i>Biotechnol. Adv.</i>, 30(5), pp. 1185-1194. <br/><br/> |
| − | Garamella, J., Marshall, R., Rustad, M. & Noireaux, V., 2016. The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. <i>ACS Syn. Biol.</i>, 5(4), pp. 344-355.<br /> | + | Garamella, J., Marshall, R., Rustad, M. & Noireaux, V., 2016. The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. <i>ACS Syn. Biol.</i>, 5(4), pp. 344-355. <br/><br/> |
| − | Jelenc, P. C. & Kurland, C. G., 1979. Nucleoside triphosphate regeneration decreases the frequency of translation errors. <i>Proc. Natl. Acad. Sci. USA</i>, 76(7), pp. 3174-3178.<br /> | + | Jelenc, P. C. & Kurland, C. G., 1979. Nucleoside triphosphate regeneration decreases the frequency of translation errors. <i>Proc. Natl. Acad. Sci. USA</i>, 76(7), pp. 3174-3178. <br/><br/> |
| − | Jewett, M. C. & Swartz, J. R., 2004. Mimicking the <i>Escherichia coli</i> cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. <i>Biotechnol. & Bioeng.</i>, 86(1), pp. 19-26.<br /> | + | Jewett, M. C. & Swartz, J. R., 2004. Mimicking the <i>Escherichia coli</i> cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. <i>Biotechnol. & Bioeng.</i>, 86(1), pp. 19-26. <br/><br/> |
| − | Jewett, M. C. et al., 2008. An integrated cell-free metabolic platform for protein production and synthetic biology. <i>Mol. Syst. Biol.</i>, 4(220).<br /> | + | Jewett, M. C. et al., 2008. An integrated cell-free metabolic platform for protein production and synthetic biology. <i>Mol. Syst. Biol.</i>, 4(220). <br/><br/> |
| − | Katsura, K. et al., 2017. A reproducible and scalable procedure for preparing bacterial extracts for cell-free protein synthesis. <i>J. Biochem.</i>, 162(5), pp. 357-369.<br /> | + | Katsura, K. et al., 2017. A reproducible and scalable procedure for preparing bacterial extracts for cell-free protein synthesis. <i>J. Biochem.</i>, 162(5), pp. 357-369. <br/><br/> |
| − | Kelwick, R., Webb, A. J., MacDonald, J. & Freemont, P. S., 2016. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. <i>Metab. Eng.</i>, Volume 38, pp. 370-381.<br /> | + | Kelwick, R., Webb, A. J., MacDonald, J. & Freemont, P. S., 2016. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. <i>Metab. Eng.</i>, Volume 38, pp. 370-381. <br/><br/> |
| − | Kwon, Y. & Jewett, M. C., 2015. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. <i>Sci. Rep.</i>, Volume 5.<br /> | + | Kwon, Y. & Jewett, M. C., 2015. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. <i>Sci. Rep.</i>, Volume 5. <br/><br/> |
| − | Lee, K. H. & Kim, D. M., 2013. Applications of cell-free protein synthesis in synthetic biology: Interfacing bio-machinery with synthetic environments. <i>Biotechnol. J.</i>, 8(11), pp. 1292-1300.<br /> | + | Lee, K. H. & Kim, D. M., 2013. Applications of cell-free protein synthesis in synthetic biology: Interfacing bio-machinery with synthetic environments. <i>Biotechnol. J.</i>, 8(11), pp. 1292-1300. <br/><br/> |
| − | Lu, Y., 2017. Cell-free synthetic biology: Engineering in an open world. <i>Syn. and Sys. Biotech.</i>, 2(1), pp. 23-27.<br /> | + | Lu, Y., 2017. Cell-free synthetic biology: Engineering in an open world. <i>Syn. and Sys. Biotech.</i>, 2(1), pp. 23-27. <br/><br/> |
| − | Nierhaus, K. H., 2014. Mg2+, K+, and the Ribosome. <i>J. Bacteriol.</i>, 196(22), pp. 3817-3819.<br /> | + | Nierhaus, K. H., 2014. Mg2+, K+, and the Ribosome. <i>J. Bacteriol.</i>, 196(22), pp. 3817-3819. <br/><br/> |
| − | Nirenberg, M. W. & Matthaei, J. H., 1961. The dependence of cell-free protein synthesis in <i>E. coli</i> upon naturally occurring or synthetic polyribonucleotides. <i>Proc. Natl. Acad. Sci. USA</i>, 47(10), pp. 1588-1602.<br /> | + | Nirenberg, M. W. & Matthaei, J. H., 1961. The dependence of cell-free protein synthesis in <i>E. coli</i> upon naturally occurring or synthetic polyribonucleotides. <i>Proc. Natl. Acad. Sci. USA</i>, 47(10), pp. 1588-1602. <br/><br/> |
| − | Pyle, A. M., 2002. Metal ions in the structure and function of RNA. <i>J. Biol. Inorg.</i>, 7(8), pp. 679-690.<br /> | + | Pyle, A. M., 2002. Metal ions in the structure and function of RNA. <i>J. Biol. Inorg.</i>, 7(8), pp. 679-690. <br/><br/> |
| − | SAS Institute Inc., 2016. JMP® 13 Design of Experiments Guide. Cary, NC, USA: SAS Institute Inc.<br /> | + | SAS Institute Inc., 2016. JMP® 13 Design of Experiments Guide. Cary, NC, USA: SAS Institute Inc. <br/><br/> |
Yang, W. C., Patel, K. & Wong, H. E., 2012. Simplifying and streamlining <i>Escherichia coli</i>-based cell-free protein synthesis. <i>Biotechnol. Prog.</i>, 28(2), pp. 413-420.<br /> | Yang, W. C., Patel, K. & Wong, H. E., 2012. Simplifying and streamlining <i>Escherichia coli</i>-based cell-free protein synthesis. <i>Biotechnol. Prog.</i>, 28(2), pp. 413-420.<br /> | ||
| Line 2,017: | Line 2,028: | ||
document.getElementById("nav-cellfree").classList.remove("active", "show"); | document.getElementById("nav-cellfree").classList.remove("active", "show"); | ||
} | } | ||
| + | |||
| + | |||
| + | $("#key_res").click(function(){ | ||
| + | $(".keyres_hide").toggle(); | ||
| + | }); | ||
Latest revision as of 22:39, 1 November 2017
spacefill
spacefill
Our Experimental Results
Key Achievements - click to show
- Demonstrated that biosensors can be successfully split into three modules
- Produced biosensor variants by co-culturing different module variants together
- Used 3D spatial modelling to begin optimisation of a multicellular biosensor
- Characterised a 'standby switch' based on an improved part (BBa_K1632007)
- Demonstrated that a Design of Experiments approach can be used to optimise cell-free systems
Below is a diagram of our Sensynova Framework. Clicking on each part of the framework (e.g. detector modules) links to the relevant results.
Alternatively, at the bottom of this page are tabs which will show you results for every part of the project
 |
 |
 |
 |
 |
 |
 |
 |


















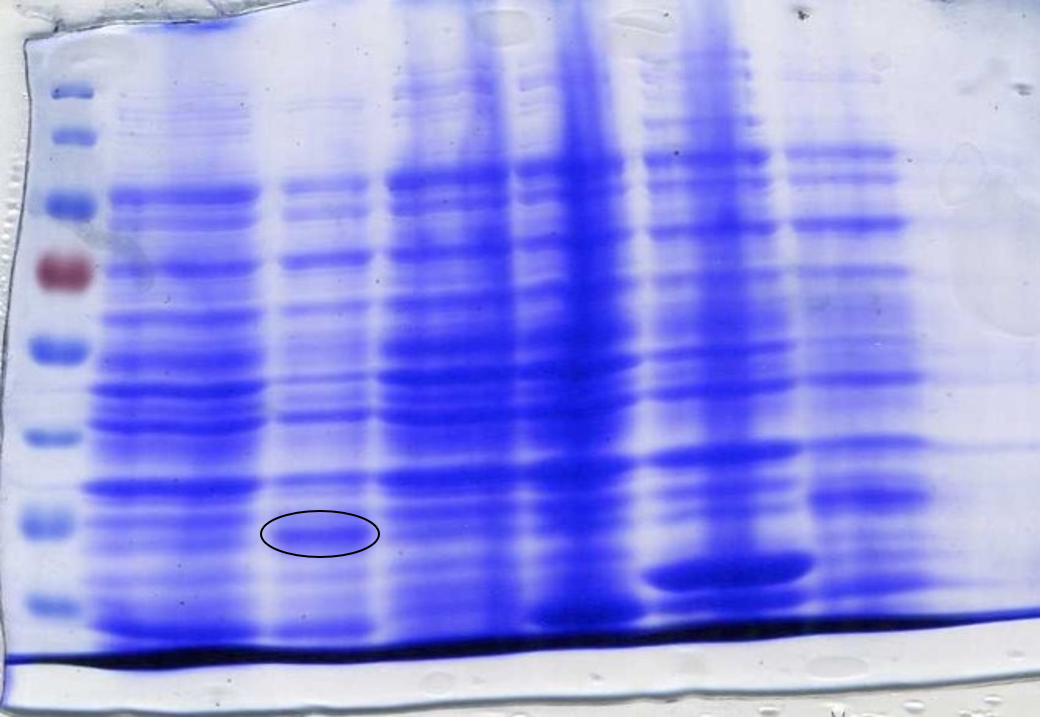












































 The
The