Caoxinyue1 (Talk | contribs) |
|||
| Line 596: | Line 596: | ||
<h4>OVERVIEW</h4> | <h4>OVERVIEW</h4> | ||
| + | <hr> | ||
<p>After doing relevant literature reading, we found that yeast’s tolerance level of ambient copper and cadmium ions has a threshold concentration, approximately 3mM and 0.5mM in SC culture media respectively.</p> | <p>After doing relevant literature reading, we found that yeast’s tolerance level of ambient copper and cadmium ions has a threshold concentration, approximately 3mM and 0.5mM in SC culture media respectively.</p> | ||
<p> In order to increase yeast strains’ inherent tolerance of copper or/and cadmium ions in their growing environment, we used this cutting-edge biological technology—SCRaMbLE, which stands for Synthetic Chromosome Rearrangement and Modification by <i>Loxpsym</i>-mediated Evolution, to obtain yeast strain with better tolerance to heavy metal ions .<sup>[1]</sup>. </p> | <p> In order to increase yeast strains’ inherent tolerance of copper or/and cadmium ions in their growing environment, we used this cutting-edge biological technology—SCRaMbLE, which stands for Synthetic Chromosome Rearrangement and Modification by <i>Loxpsym</i>-mediated Evolution, to obtain yeast strain with better tolerance to heavy metal ions .<sup>[1]</sup>. </p> | ||
| Line 601: | Line 602: | ||
<p>In order to characterize their increased tolerance of copper or/and cadmium ions, we designed and conducted two different sets of experiments, in both visible and quantitative manner, to test their ability to cope with adverse environmental conditions.</p> | <p>In order to characterize their increased tolerance of copper or/and cadmium ions, we designed and conducted two different sets of experiments, in both visible and quantitative manner, to test their ability to cope with adverse environmental conditions.</p> | ||
<h4>CONSTRUCTION</h4> | <h4>CONSTRUCTION</h4> | ||
| + | <hr> | ||
<p>This vector consists of three parts, an estrogen-inducible PCLB2 promoter <a href="http://parts.igem.org/Part:BBa_K2407002">(BBa_K2407002)</a>, the Cre-EBD sequence <a href="http://parts.igem.org/Part:BBa_K2407005">(BBa_K2407005)</a> and a CYC1 terminator<a href="http://parts.igem.org/Part:BBa_K2407003">(BBa_K2407003)</a>. We used overlap PCR to ligate these three parts and then the plasmids with URA3 and HIS nutritional label respectively through enzymatic digestion and ligation. Then this composite part <a href="http://parts.igem.org/Part:BBa_K2407011">(BBa_K2407011)</a>,was sequenced and proved to be accurate by using the promoter's forward primer and the terminator's reverse primer. The electrophoresis results below also showcased that the sequence length (about 2800bp) was correct.</p> | <p>This vector consists of three parts, an estrogen-inducible PCLB2 promoter <a href="http://parts.igem.org/Part:BBa_K2407002">(BBa_K2407002)</a>, the Cre-EBD sequence <a href="http://parts.igem.org/Part:BBa_K2407005">(BBa_K2407005)</a> and a CYC1 terminator<a href="http://parts.igem.org/Part:BBa_K2407003">(BBa_K2407003)</a>. We used overlap PCR to ligate these three parts and then the plasmids with URA3 and HIS nutritional label respectively through enzymatic digestion and ligation. Then this composite part <a href="http://parts.igem.org/Part:BBa_K2407011">(BBa_K2407011)</a>,was sequenced and proved to be accurate by using the promoter's forward primer and the terminator's reverse primer. The electrophoresis results below also showcased that the sequence length (about 2800bp) was correct.</p> | ||
| Line 617: | Line 619: | ||
<h4>CHARACTERIZATION</h4> | <h4>CHARACTERIZATION</h4> | ||
| + | <hr> | ||
<h5>Dilution Assay </h5> | <h5>Dilution Assay </h5> | ||
<p>We conducted dilution assay on SC solid media containing 0.14 mM cadmium ions. Experimental groups are S1, S2, S3, and S4; control groups are <i>synX</i> (the yeast strain containing a synthetic chromosome X), <i>BY4741</i> (wild-type haploid yeast), and <i>BY4743</i> (wild-type diploid yeast). Results are shown in the picture below.</p> | <p>We conducted dilution assay on SC solid media containing 0.14 mM cadmium ions. Experimental groups are S1, S2, S3, and S4; control groups are <i>synX</i> (the yeast strain containing a synthetic chromosome X), <i>BY4741</i> (wild-type haploid yeast), and <i>BY4743</i> (wild-type diploid yeast). Results are shown in the picture below.</p> | ||
| Line 705: | Line 708: | ||
<p>The Fig.3-6 and Fig.3-7 showcase that the survival rate of S5 is higher than that of <i>synX</i> after yeast cells are immersed in copper ions solutions of identical concentration for the same amount of time. The quantitative results are that compared with the control strain, the experimental strain S5's ability to tolerate copper ions has increased by 74% (1 hour), 72% (2 hours), and 698% (3 hours). It also can be extrapolated that the gap of survival rates between the mutated strain and <i>synX</i> strain will continue to widen as the immersion time increases. The results are consistent with the dilution assay too. </p> | <p>The Fig.3-6 and Fig.3-7 showcase that the survival rate of S5 is higher than that of <i>synX</i> after yeast cells are immersed in copper ions solutions of identical concentration for the same amount of time. The quantitative results are that compared with the control strain, the experimental strain S5's ability to tolerate copper ions has increased by 74% (1 hour), 72% (2 hours), and 698% (3 hours). It also can be extrapolated that the gap of survival rates between the mutated strain and <i>synX</i> strain will continue to widen as the immersion time increases. The results are consistent with the dilution assay too. </p> | ||
<h4>EXPECTATIONS</h4> | <h4>EXPECTATIONS</h4> | ||
| + | <hr> | ||
<p>We are exhilarated to see that SCRaMbLE is really a feasible technology to enhance the yeast's ability to cope with adverse environmental conditions. Not just heavy metal ions, we are looking forward to seeing its future applications, be they, for example, alcohol tolerance or heat tolerance. </p> | <p>We are exhilarated to see that SCRaMbLE is really a feasible technology to enhance the yeast's ability to cope with adverse environmental conditions. Not just heavy metal ions, we are looking forward to seeing its future applications, be they, for example, alcohol tolerance or heat tolerance. </p> | ||
| Line 1,079: | Line 1,083: | ||
<h4>Accumulation</h4> | <h4>Accumulation</h4> | ||
| + | <hr> | ||
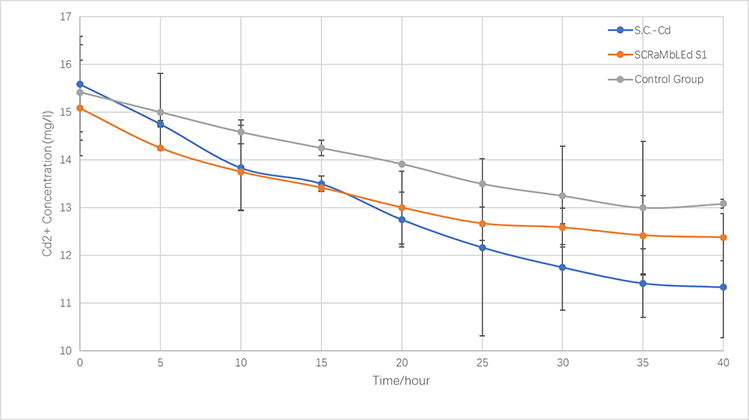
<p>Firstly, <i>S.C-Cu</i>, <i>S8</i> (screened by SCRaMbLE) and <i>BY4741</i> are cultured in YPD liquid media for 24 hours. Then add the 430 mg/L copper ions solution. Cells are cultured for another 45 hours (30℃). Atomic absorption spectroscopy is used to measure the concentration of copper ions in the supernatant every 5 hours. We depict the adsorption curve of copper ions.</p> | <p>Firstly, <i>S.C-Cu</i>, <i>S8</i> (screened by SCRaMbLE) and <i>BY4741</i> are cultured in YPD liquid media for 24 hours. Then add the 430 mg/L copper ions solution. Cells are cultured for another 45 hours (30℃). Atomic absorption spectroscopy is used to measure the concentration of copper ions in the supernatant every 5 hours. We depict the adsorption curve of copper ions.</p> | ||
<p>In the same way, we culture Cd Yeast, <i>S1</i> (screened by SCRaMbLE) and <i>BY4741</i> in YPD liquid medium for 24 hours and then add xx mg/L cadmium ions solution to the media for another xx hours (30℃). the concentration of cadmium ions in the supernatant is measured every x hours. | <p>In the same way, we culture Cd Yeast, <i>S1</i> (screened by SCRaMbLE) and <i>BY4741</i> in YPD liquid medium for 24 hours and then add xx mg/L cadmium ions solution to the media for another xx hours (30℃). the concentration of cadmium ions in the supernatant is measured every x hours. | ||
| Line 1,119: | Line 1,124: | ||
<p>Moreover, it’s a pity that we have no time to combine our genetic circuit and SCRaMbLE yeast. In subsequent experiments, it deserves a try to transform fragments into SCRaMbLE yeast, checking if it will double the absorption capacity or even better.</p> | <p>Moreover, it’s a pity that we have no time to combine our genetic circuit and SCRaMbLE yeast. In subsequent experiments, it deserves a try to transform fragments into SCRaMbLE yeast, checking if it will double the absorption capacity or even better.</p> | ||
<p>In the nutshell, we will be dedicated to improve the absorption efficiency and better our genetic circuit.</p> | <p>In the nutshell, we will be dedicated to improve the absorption efficiency and better our genetic circuit.</p> | ||
| + | <div class="reference"> | ||
<h4>Reference</h4> | <h4>Reference</h4> | ||
<hr> | <hr> | ||
| Line 1,125: | Line 1,131: | ||
<p>[2]C. Baumann, A. Beil, S. Jurt, M. Niederwanger, O. Palacios, M. Capdevila, S. Atrian, R. Dallinger, O. Zerbe, Angew. Chem. Int. Ed. 2017, 56, 4617.</p> | <p>[2]C. Baumann, A. Beil, S. Jurt, M. Niederwanger, O. Palacios, M. Capdevila, S. Atrian, R. Dallinger, O. Zerbe, Angew. Chem. Int. Ed. 2017, 56, 4617.</p> | ||
<p>[3]Dönmez G, Aksu Z. The effect of copper(II) ions on the growth and bioaccumulation properties of some yeasts[J]. Process Biochemistry, 1999, 35(1–2):135-142.</p> | <p>[3]Dönmez G, Aksu Z. The effect of copper(II) ions on the growth and bioaccumulation properties of some yeasts[J]. Process Biochemistry, 1999, 35(1–2):135-142.</p> | ||
| − | + | </div> | |
<div class="collapse-card__close_handler mt1 align-right mouse-pointer"> | <div class="collapse-card__close_handler mt1 align-right mouse-pointer"> | ||
Revision as of 12:19, 1 November 2017
/* OVERRIDE IGEM SETTINGS */