Caoxinyue1 (Talk | contribs) |
|||
| Line 1,077: | Line 1,077: | ||
<a href="#pic_seventy-five"> | <a href="#pic_seventy-five"> | ||
<img src="https://static.igem.org/mediawiki/2017/b/b7/Heavy-metal-jiaotu.jpg"></a> | <img src="https://static.igem.org/mediawiki/2017/b/b7/Heavy-metal-jiaotu.jpg"></a> | ||
| − | <p style="font-size:15px;text-align:center"><br/>Figure 5-1. The results of PCR of our S.C-Cu. LIMT gene (length of 319bp) | + | <p style="font-size:15px;text-align:center"><br/>Figure 5-1. The results of PCR of our <i>S.C-Cu</i>. <i>LIMT</i> gene (length of 319bp) 、<i>Cup1</i>(length of 186bp) and complete sequence(length of 3114bp)have been amplified. which indicated that we succeeded in the construction of genetic circuit.</p> |
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="pic_seventy-five" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/b/b7/Heavy-metal-jiaotu.jpg"><p style="font-size:15px;text-align:center"><br/>Figure 5-1. The results of PCR of our S.C-Cu. LIMT gene (length of 319bp) | + | <div id="pic_seventy-five" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/b/b7/Heavy-metal-jiaotu.jpg"><p style="font-size:15px;text-align:center"><br/>Figure 5-1. The results of PCR of our <i>S.C-Cu</i>. <i>LIMT</i> gene (length of 319bp) 、<i>Cup1</i>(length of 186bp) and complete sequence(length of 3114bp)have been amplified. which indicated that we succeeded in the construction of genetic circuit.</p></div> |
<p>Fig.5-1 the results of PCR. We use <i>2k plus Ⅱ</i> as the marker. On four parallel lanes of the gel (number 1,2,3,4), run were four set of DNA molecules of known size (319bp for number 1, the <i>LIMT</i>; 186bp for number 2 and 3, the <i>Cup1</i>; 3114bp for number 4,the whole sequence contained <i>Cup1</i>). From the DNA band of number 1, we could analyze that <i>vika</i> has been expressed to delete the <i>Cup1</i> and its terminor, so we can get the <i>LIMT</i>. From the DNA band of number 2, 3 and 4, we could delightedly prove that the fragments (<i>TEF</i> promoter, <i>Cup1</i> and <i>ura3</i> terminator) have successfully transformed to synthetic chromosome <i>V</i>. </p> | <p>Fig.5-1 the results of PCR. We use <i>2k plus Ⅱ</i> as the marker. On four parallel lanes of the gel (number 1,2,3,4), run were four set of DNA molecules of known size (319bp for number 1, the <i>LIMT</i>; 186bp for number 2 and 3, the <i>Cup1</i>; 3114bp for number 4,the whole sequence contained <i>Cup1</i>). From the DNA band of number 1, we could analyze that <i>vika</i> has been expressed to delete the <i>Cup1</i> and its terminor, so we can get the <i>LIMT</i>. From the DNA band of number 2, 3 and 4, we could delightedly prove that the fragments (<i>TEF</i> promoter, <i>Cup1</i> and <i>ura3</i> terminator) have successfully transformed to synthetic chromosome <i>V</i>. </p> | ||
| Line 1,096: | Line 1,096: | ||
<div class="small_pic_demo_qqqqqqq" style="float:left;"> | <div class="small_pic_demo_qqqqqqq" style="float:left;"> | ||
<a href="#pic_Seventy-six"> | <a href="#pic_Seventy-six"> | ||
| − | <img src="https://static.igem.org/mediawiki/parts/1/17/Demonstrate.Cu.png"></a><p style="font-size:15px;text-align:center"><br/>Figure 5-2.The variations of copper(II) consumption with time for S.C- | + | <img src="https://static.igem.org/mediawiki/parts/1/17/Demonstrate.Cu.png"></a><p style="font-size:15px;text-align:center"><br/>Figure 5-2.The variations of copper(II) consumption with time for <i>S.C-Cu</i>、<i>S8</i> and <i>BY4741</i> at 430 mg/L copper(II) concentrations. |
</p> | </p> | ||
</div> | </div> | ||
| Line 1,102: | Line 1,102: | ||
<a href="#pic_Seventy-seven" > | <a href="#pic_Seventy-seven" > | ||
<img src="https://static.igem.org/mediawiki/parts/5/5f/Demonstrate.Cd.png"/> | <img src="https://static.igem.org/mediawiki/parts/5/5f/Demonstrate.Cd.png"/> | ||
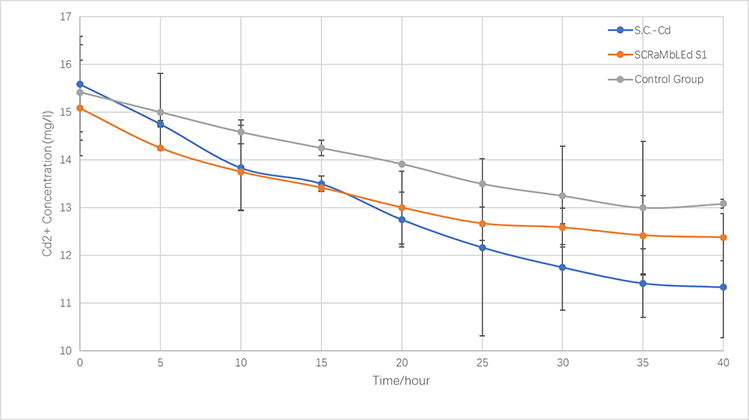
| − | </a> <p style="font-size:15px;text-align:center"><br/>Figure 5-3.The variations of cadmium(II) consumption with time for S.C- | + | </a> <p style="font-size:15px;text-align:center"><br/>Figure 5-3.The variations of cadmium(II) consumption with time for <i>S.C-Cd</i>、<i>S1</i> and <i>BY4741</i> at 16 mg/L cadmium(II) concentrations</p> |
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="pic_Seventy-six" style="display:none;"><img src="https://static.igem.org/mediawiki/parts/1/17/Demonstrate.Cu.png"/><p style="font-size:15px;text-align:center"><br/>The variations of copper(II) consumption with time for S.C- | + | <div id="pic_Seventy-six" style="display:none;"><img src="https://static.igem.org/mediawiki/parts/1/17/Demonstrate.Cu.png"/><p style="font-size:15px;text-align:center"><br/>The variations of copper(II) consumption with time for <i>S.C-Cu</i>、<i>S8</i> and <i>BY4741</i> at 430 mg/L copper(II) concentrations. |
</p></div> | </p></div> | ||
| − | <div id="pic_Seventy-seven" style="display:none;"><img src="https://static.igem.org/mediawiki/parts/5/5f/Demonstrate.Cd.png"/><p style="font-size:15px;text-align:center"><br/>Figure 5-3.The variations of cadmium(II) consumption with time for S.C- | + | <div id="pic_Seventy-seven" style="display:none;"><img src="https://static.igem.org/mediawiki/parts/5/5f/Demonstrate.Cd.png"/><p style="font-size:15px;text-align:center"><br/>Figure 5-3.The variations of cadmium(II) consumption with time for <i>S.C-Cd</i>、<i>S1</i> and <i>BY4741</i> at 16 mg/L cadmium(II) concentrations </p></div> |
<p>Afterwards, we check if the <i>vika</i> enzyme could work well. The Cu yeast with a plasmid expressing <i>vika</i> enzyme is grew in the medium with <i> raffinose</i>, then transferred to heavy metal solution. | <p>Afterwards, we check if the <i>vika</i> enzyme could work well. The Cu yeast with a plasmid expressing <i>vika</i> enzyme is grew in the medium with <i> raffinose</i>, then transferred to heavy metal solution. | ||
<div class="zxx_zoom_demo_qqqqqqq" align="center"> | <div class="zxx_zoom_demo_qqqqqqq" align="center"> | ||
| Line 1,115: | Line 1,115: | ||
<a href="#pic_eighty"> | <a href="#pic_eighty"> | ||
<img src="https://static.igem.org/mediawiki/2017/6/6b/Design.cu-cd.curve.png"></a> | <img src="https://static.igem.org/mediawiki/2017/6/6b/Design.cu-cd.curve.png"></a> | ||
| − | <p style="font-size:15px;text-align:center"><br/>Figure 5-4. S.C-Cu is cultivated in medium with <i> raffinose </i> including 320mg/L copper ions and 6mg/L Cadmium.<i>galactose</i> is added at 12 hours to turn on "switch".</p> | + | <p style="font-size:15px;text-align:center"><br/>Figure 5-4. <i>S.C-Cu</i> is cultivated in medium with <i> raffinose </i> including 320mg/L copper ions and 6mg/L Cadmium.<i>galactose</i> is added at 12 hours to turn on "switch".</p> |
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="pic_eighty" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/6/6b/Design.cu-cd.curve.png"><p style="font-size:15px;text-align:center"><br/>Figure 5-4. S.C-Cu is cultivated in medium with <i> raffinose </i> including 320mg/L copper ions and 6mg/L Cadmium.<i>galactose</i> is added at 12 hours to turn on "switch".</p></div> | + | <div id="pic_eighty" style="display:none;"><img src="https://static.igem.org/mediawiki/2017/6/6b/Design.cu-cd.curve.png"><p style="font-size:15px;text-align:center"><br/>Figure 5-4. <i>S.C-Cu</i> is cultivated in medium with <i> raffinose </i> including 320mg/L copper ions and 6mg/L Cadmium.<i>galactose</i> is added at 12 hours to turn on "switch".</p></div> |
<p>Fig.5-4 clearly shows the change of the concentration of heavy metal ions in the supernatant. Firstly, the Cu yeast works smoothly. The concentration of copper ions declines over time while that of cadmium ions barely changes. 12 hours later, we add <i>galactose</i> to the solution. Situation changes. <i>Galactose</i> induces the enzyme, changing Cu yeast to Cd yeast. It leads to faster adsorption of cadmium but slower for copper.</p> | <p>Fig.5-4 clearly shows the change of the concentration of heavy metal ions in the supernatant. Firstly, the Cu yeast works smoothly. The concentration of copper ions declines over time while that of cadmium ions barely changes. 12 hours later, we add <i>galactose</i> to the solution. Situation changes. <i>Galactose</i> induces the enzyme, changing Cu yeast to Cd yeast. It leads to faster adsorption of cadmium but slower for copper.</p> | ||
Revision as of 17:13, 1 November 2017
/* OVERRIDE IGEM SETTINGS */