Caoxinyue1 (Talk | contribs) |
|||
| Line 127: | Line 127: | ||
margin:0.5em; | margin:0.5em; | ||
font-family: 'LiberationSerif-Bold',Verdana,sans-serif; | font-family: 'LiberationSerif-Bold',Verdana,sans-serif; | ||
| + | text-transform:uppercase; | ||
| + | |||
} | } | ||
#normal h5{ | #normal h5{ | ||
| Line 458: | Line 460: | ||
</p> | </p> | ||
<h4>EXPECTED RESULTS</h4> | <h4>EXPECTED RESULTS</h4> | ||
| + | <hr> | ||
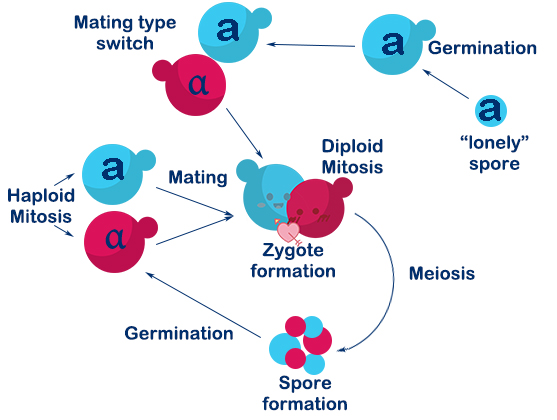
<p>In our design, Mating Switcher is a means of gene regulation. We can transform from one functional system to another system through this switch conveniently. To show the function of Mating Switcher more intuitively, we construct this<i> RFP</i> system to be a characterization. | <p>In our design, Mating Switcher is a means of gene regulation. We can transform from one functional system to another system through this switch conveniently. To show the function of Mating Switcher more intuitively, we construct this<i> RFP</i> system to be a characterization. | ||
| Line 513: | Line 516: | ||
<div class="collapse-card__body"> | <div class="collapse-card__body"> | ||
<h4>Overview</h4> | <h4>Overview</h4> | ||
| + | <hr> | ||
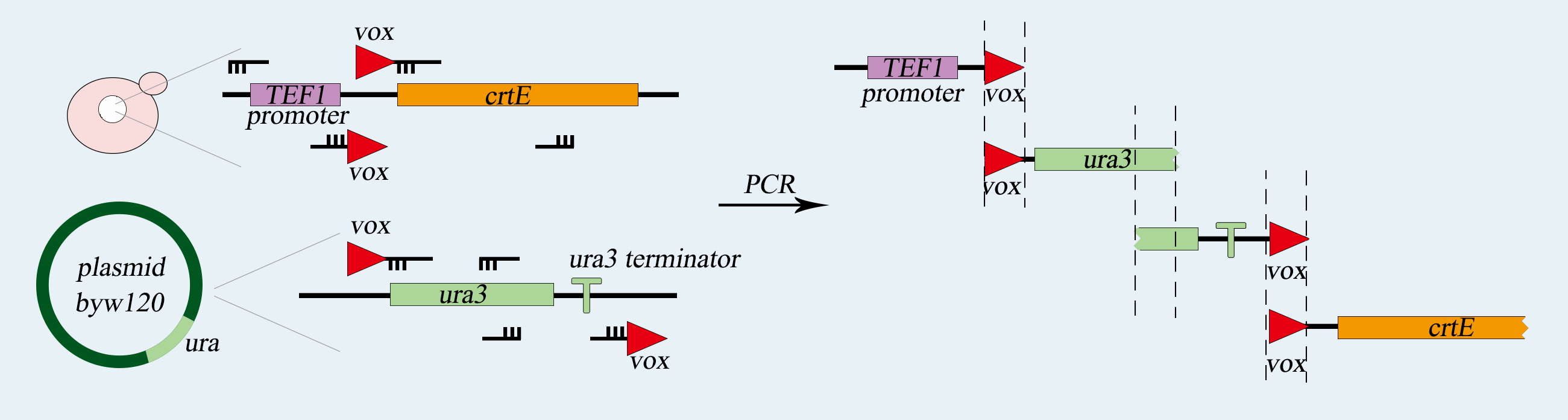
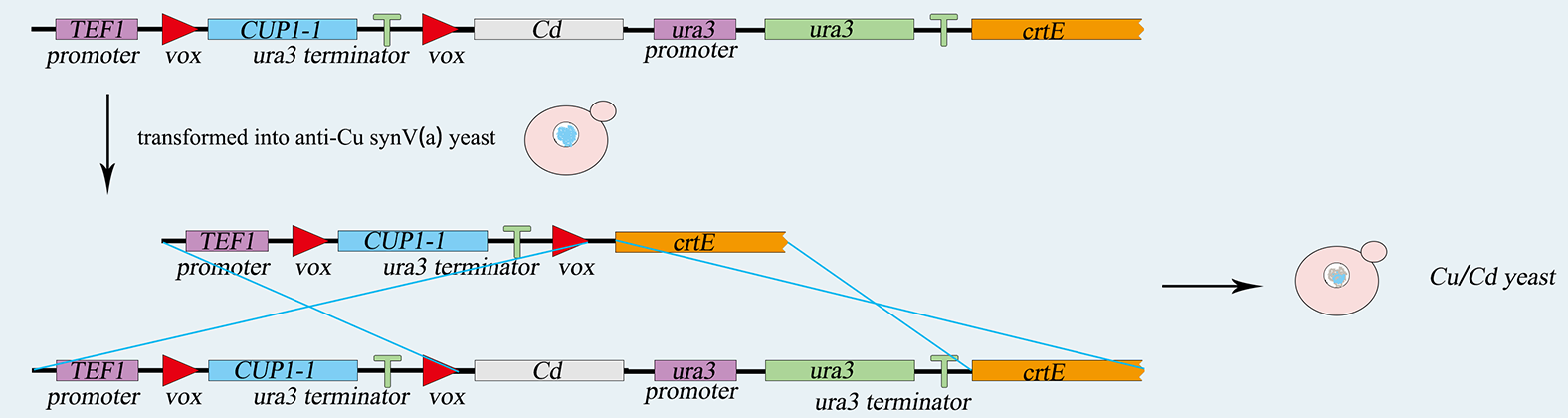
<p>Synthetic chromosome rearrangement and modification by <i>loxP</i>-mediated evolution(SCRaMble) generates combinatorial geomic diversity through rearrangements at designed recombinase sites. We applied SCRaMble to Saccharomyces cerevisiae(<i>synX</i>)to attain strains with better tolerance to high concentration of cadmium ion and cupric ion solution and compared the growing condition with the original strains to demonstrate the validity of SCRaMble.</p> | <p>Synthetic chromosome rearrangement and modification by <i>loxP</i>-mediated evolution(SCRaMble) generates combinatorial geomic diversity through rearrangements at designed recombinase sites. We applied SCRaMble to Saccharomyces cerevisiae(<i>synX</i>)to attain strains with better tolerance to high concentration of cadmium ion and cupric ion solution and compared the growing condition with the original strains to demonstrate the validity of SCRaMble.</p> | ||
<h4>Theoretical Background</h4> | <h4>Theoretical Background</h4> | ||
| + | <hr> | ||
<h5>Sc2.0 Project</h5> | <h5>Sc2.0 Project</h5> | ||
<p>Synthetic Yeast Genome Project (Sc2.0) is the world’s first synthetic eukaryotic genome project that aims to create a novel, rationalized version of the genome of the yeast species Saccharomyces cerevisiae. On March 10th, 7 articles related to Sc 2.0 were published on Science. As a member of Sc 2.0, YJ lab has completed two synthetic yeast chromosomes, and two articles are published on Science discussing challenging but exciting task of building synthetic chromosomes V, X .</p> | <p>Synthetic Yeast Genome Project (Sc2.0) is the world’s first synthetic eukaryotic genome project that aims to create a novel, rationalized version of the genome of the yeast species Saccharomyces cerevisiae. On March 10th, 7 articles related to Sc 2.0 were published on Science. As a member of Sc 2.0, YJ lab has completed two synthetic yeast chromosomes, and two articles are published on Science discussing challenging but exciting task of building synthetic chromosomes V, X .</p> | ||
| Line 536: | Line 541: | ||
<h4>Experiment Design</h4> | <h4>Experiment Design</h4> | ||
| + | <hr> | ||
<h5>Construction of Cre-<i>loxpsym</i> System </h5> | <h5>Construction of Cre-<i>loxpsym</i> System </h5> | ||
<p>First, We use two common expression vector plasmid, PRS416 and PRS413(with different nutrition label), in Saccharomyces cerevisiae to load our device, which consists of heterologous gene part (PCLB2 promoter, Cre-EBD gene, CYC1 terminator) . Second, we transform the pSCW11-CRE/EBD plasmid into <i>synX</i> strain , respectively and get three strains with Cre-EBD, 079 and 160 with ura tag ,085 with his tag.</p> | <p>First, We use two common expression vector plasmid, PRS416 and PRS413(with different nutrition label), in Saccharomyces cerevisiae to load our device, which consists of heterologous gene part (PCLB2 promoter, Cre-EBD gene, CYC1 terminator) . Second, we transform the pSCW11-CRE/EBD plasmid into <i>synX</i> strain , respectively and get three strains with Cre-EBD, 079 and 160 with ura tag ,085 with his tag.</p> | ||
Revision as of 12:15, 1 November 2017
/* OVERRIDE IGEM SETTINGS */
Design
Background
Human existence on earth is almost impossible without the heavy metals. Even though important to mankind, exposure to them during production, usage and their uncontrolled discharge into the environment has caused lots of hazards to man, other organisms and the environment itself. Heavy metals can enter human tissues and organs via inhalation, diet, and manual handling. As the process of urbanization and industrialization goes deeper and deeper, heavy metal pollution, a noticeable threaten to almost all the creatures, has become an essential problem to solve.
According to our human practice, the situation of heavy metal pollution (copper and cadmium ions) is marked on a world map, and the severity of heavy metal pollution has been increasing all over this map. Places with serious pollution include middle Asia, eastern Asia, southern Europe, and Latin America. In addition, not only fresh water source, but also soil and crops are seriously contaminated by heavy metals. On average, during three out of ten suppers we have, we absorb excess heavy metals over the standard concentration.
Considering the rigorous situation we face, our team decided to design an advanced system for typical toxic heavy metal disposal based on Saccharomyces cerevisiae.