| (16 intermediate revisions by 3 users not shown) | |||
| Line 23: | Line 23: | ||
padding-right: 60px; | padding-right: 60px; | ||
} | } | ||
| − | .fig>figure{ | + | |
| + | .fig>figure { | ||
padding-bottom: 20px; | padding-bottom: 20px; | ||
padding-top: 20px; | padding-top: 20px; | ||
| Line 37: | Line 38: | ||
<div class="container"> | <div class="container"> | ||
<div class="para"> | <div class="para"> | ||
| − | <h1> | + | <p style="font-size: 25px"> |
| + | The peptide production contains two parts: cross-inhibition and antimicrobial peptides | ||
| + | </p> | ||
| + | <h1>Cross-inhibition</h1> | ||
| + | |||
| + | |||
| + | <p style="font-size:20px"> | ||
| + | As introduced in the description section, there are four different classes in the Agr quorum sensing systems which are referred | ||
| + | to as Agr-I, Agr-II, Agr-III, and Agr-IV and each is capable of recognizing a unique AIP structure, which | ||
| + | is AIP-I, AIP-II, AIP-III and AIP-IV, respectively. | ||
| + | <i>S. aureus </i>may also be classified into four groups/strains (I to IV) according to the class of the AIPs | ||
| + | produced. Similarly, the proteins involved in signal biosynthesis (AgrB and AgrD) and surface receptor binding | ||
| + | (AgrC) also show variability. Interestingly, different AIP signals cross-inhibit the activity of the others. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="fig" align="center"> | ||
| + | <figure> | ||
| + | <img width="650px" src="https://static.igem.org/mediawiki/2017/9/93/Detection2.png"> | ||
| + | <figcaption> | ||
| + | <strong>Figure 1a The structure and cross-inhibition of AIPs</strong> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <p style="font-size:20px">For the cross-inhibition, we introduce two genes, agrB and agrD from a different type of | ||
| + | <i>S.aureus</i>. The transmembrane protein AgrB cleaves the precursor peptide AgrB into a different type of | ||
| + | AIPs which is capable of inhibiting the combination of the original AIPs with the membrane AgrC, and thus | ||
| + | stopping the quorum sensing and growth of biofilms. The construction is shown below.</p> | ||
| + | </div> | ||
| + | |||
| + | <div class="fig" align="center"> | ||
| + | <figure> | ||
| + | <img width="650px" src="https://static.igem.org/mediawiki/2017/0/09/%E5%BE%AE%E4%BF%A1%E6%88%AA%E5%9B%BE_20171116173734.jpg"> | ||
| + | <figcaption> | ||
| + | <strong>Figure 1b The construction of the cross-inhibition part</strong> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <h1>Antimicrobial Peptide</h1> | ||
| + | <img class="img-responsive center-block" src=" https://static.igem.org/mediawiki/2017/3/3f/Circuit_Peptides.png " height=700 width=800> | ||
<p style="font-size:20px"> | <p style="font-size:20px"> | ||
Anti-microbial peptide (AMP) is a part of the innate immune system of most multi-cellular organisms to counter microbial | Anti-microbial peptide (AMP) is a part of the innate immune system of most multi-cellular organisms to counter microbial | ||
| − | infections (Margitta and Torsten, 1999). The cationic and amphipathic α- | + | infections (Margitta and Torsten, 1999). Among these structural groups, α-helix and β-sheet structures are |
| − | + | more common. AMPs kill cells by disrupting membrane integrity (via interaction with the negatively charged | |
| − | + | molecules on the cell membrane), by inhibiting proteins, DNA and RNA synthesis, or by interacting with certain | |
| − | + | intracellular targets. All AMPs known by the late-90s are cationic. However, the concept that AMPs need to | |
| + | be cationic was changed later with the discovery of negatively charged AMPs in 1997. For example maximin-H5 | ||
| + | from frog skin and dermicidin secreted from the sweat gland tissues of humans are both anionic peptides (Bahar, | ||
| + | and Ren, 2013). | ||
| + | </p> | ||
| + | <p style="font-size:20px"> | ||
| + | One of important features of AMPs is their rapid killing effect. AMPs can influence cell membrane in few seconds after contact. | ||
| + | AMPs are also known to enhance the activities of antibiotics through their synergistic effects. For example, | ||
| + | the combination of penicillin with pediocin and ampicillin with nisin Z exhibited a killing effect on | ||
| + | <i>Pseudomonas fluorescens</i> with 13- and 155-fold lower in the minimum inhibitory concentration (MIC), respectively, | ||
| + | compared to using the antibiotics alone (Bahar, and Ren, 2013). | ||
| + | </p> | ||
| + | <p style="font-size:20px"> | ||
| + | Another feature of AMPs is their activation on the immune response defense. As shown by Figure 1c, the role of cationic host-defense | ||
| + | peptides in modulating the innate immune response and boosting infection-resolving immunity while dampening | ||
| + | potentially harmful pro-inflammatory responses gives these peptides the potential to become an entirely new | ||
| + | therapeutic approach against bacterial infections(Hancock & Sahl, 2006). | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <div class="fig" align="center"> | ||
| + | <figure> | ||
| + | <img width="650px" src="https://static.igem.org/mediawiki/2017/3/3d/The_general_function_of_antimicrobial_peptide.png"> | ||
| + | <figcaption> | ||
| + | <strong>Figure 1c: The general function of antimicrobial peptide. Antimicrobial peptides can active immune response | ||
| + | of human and kill target directly (Hancock & Sahl, 2006).</strong> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <p style="font-size:20px"> | ||
| + | Although bacteria have diverse mechanisms of resistance to AMPs, it is encouraging to notice that the general lipid bilayer | ||
| + | structure of bacterial membranes makes it hard to develop a complete resistance against AMPs. Also, the resistance | ||
| + | against AMPs reported to date is not as strong as those against antibiotics and it is only found in a limited | ||
| + | number of AMPs. | ||
| + | </p> | ||
| + | |||
| + | <h1>Cationic and amphipathic antimicrobial Peptides</h1> | ||
| + | <p style="font-size:20px"> | ||
| + | The cationic and amphipathic α-helical structure is the most common conformation in these types of peptides but some hydrophobic | ||
| + | α-helical peptides can also possess antimicrobial activity. In this project we choose three different cationic | ||
| + | antimicrobial peptides with α-helical conformation. | ||
</p> | </p> | ||
<p style="font-size:20px"> | <p style="font-size:20px"> | ||
| − | Figure | + | Figure 1d shows the molecular mechanism of cationic AMPs containing α-helical structure. Most of the cationic AMPs associate |
| − | + | with the charged lipid group of the bacteria plasma membrane. The α-helical structure disrupts the packing | |
| − | + | of lipid molecules such that the membrane becomes leaky (Rocca et al.,1999). | |
</p> | </p> | ||
</div> | </div> | ||
| Line 55: | Line 142: | ||
<figure> | <figure> | ||
<img width="650px" src="https://static.igem.org/mediawiki/2017/9/99/Peptide_Production_1.png"> | <img width="650px" src="https://static.igem.org/mediawiki/2017/9/99/Peptide_Production_1.png"> | ||
| − | <figcaption><strong> | + | <figcaption> |
| − | + | <strong>Figure 1d:The interaction mechanism of the cationic α-helical structure of anti-microbial peptides with | |
| − | + | the bacterial membrane. The α-helical structure partitions between the lipid bilayer and the aqueous | |
| + | solution. Following insertion of the peptide, the bilayer membrane permeability may be perturbed.</strong> | ||
| + | </figcaption> | ||
</figure> | </figure> | ||
</div> | </div> | ||
| Line 68: | Line 157: | ||
The peptide is cleaved from a larger protein, hCAP-18 by extracellular proteolysis of proteinase 3 from the | The peptide is cleaved from a larger protein, hCAP-18 by extracellular proteolysis of proteinase 3 from the | ||
C-terminal end of hCAP18 (Patricia, 2010; Ramos, Domingues, and Gama, 2011). The peptide composed of two | C-terminal end of hCAP18 (Patricia, 2010; Ramos, Domingues, and Gama, 2011). The peptide composed of two | ||
| − | mainly parts: from residue Leu2 to Leu31 is α-helical structure (Fig 2b) and 6 residues form | + | mainly parts: from residue Leu2 to Leu31, which is a α-helical structure (Fig 2b) and a 6 residues form the |
| − | (Fig 2a). | + | loop structure at the terminus(Fig 2a). |
</p> | </p> | ||
<p style="font-size:20px"> | <p style="font-size:20px"> | ||
Ramos, Domingues, and Gama (2011) also reported that LL-37 has additional roles such as regulating the inflammatory response | Ramos, Domingues, and Gama (2011) also reported that LL-37 has additional roles such as regulating the inflammatory response | ||
| − | + | in wounds or infection sites, binding and neutralizing LPS, and wound closure, apart from anti-microbial | |
| − | (Figure 2c). | + | property (Figure 2c). |
</p> | </p> | ||
</div> | </div> | ||
| Line 81: | Line 170: | ||
<figure> | <figure> | ||
<img width="700px" src="https://static.igem.org/mediawiki/2017/d/de/Peptide_Production_2a.png"> | <img width="700px" src="https://static.igem.org/mediawiki/2017/d/de/Peptide_Production_2a.png"> | ||
| − | <figcaption><strong> 2a: LL-37 structure and residues(PDB 2K6O)</strong></figcaption> | + | <figcaption> |
| + | <strong>Figure 2a: LL-37 structure and its amino acid residues(PDB 2K6O)</strong> | ||
| + | </figcaption> | ||
</figure> | </figure> | ||
<figure> | <figure> | ||
<img width="700px" src="https://static.igem.org/mediawiki/2017/2/29/Peptide_Production_2b.png"> | <img width="700px" src="https://static.igem.org/mediawiki/2017/2/29/Peptide_Production_2b.png"> | ||
| − | <figcaption><strong>Figure 2b: LL-37 secondary structure | + | <figcaption> |
| − | + | <strong>Figure 2b: Prediction on the LL-37 secondary structure | |
| − | + | </strong> | |
| + | </figcaption> | ||
</figure> | </figure> | ||
<figure> | <figure> | ||
| − | + | <img width="700px" src="https://static.igem.org/mediawiki/2017/3/3b/Peptide_Production_2c.png"> | |
| − | + | <figcaption> | |
| − | + | <strong> 2c: Biological functions of LL-37 (Ramos, Domingues, and Gama, 2011)</strong> | |
| + | </figcaption> | ||
| + | </figure> | ||
</div> | </div> | ||
<div class="para"> | <div class="para"> | ||
<h1>GF-17</h1> | <h1>GF-17</h1> | ||
| − | <p style="font-size:20px">GF-17 is a | + | <p style="font-size:20px">GF-17 is a highly efficient anti-microbial peptide derived from the residure Phe-17 to Val-32 of LL-37(Fig 3a). |
</p> | </p> | ||
</div> | </div> | ||
| Line 111: | Line 205: | ||
<img width="700px" src="https://static.igem.org/mediawiki/2017/8/83/Peptide_Production_3b.png"> | <img width="700px" src="https://static.igem.org/mediawiki/2017/8/83/Peptide_Production_3b.png"> | ||
<figcaption> | <figcaption> | ||
| − | <strong>Figure3b: Cartoon view of GF-17 | + | <strong>Figure3b: Cartoon view of the α-helical structure of GF-17 (PDB: 2L5M)</strong> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| Line 117: | Line 211: | ||
<div class="para"> | <div class="para"> | ||
| − | <p style="font-size:20px"> | + | <p style="font-size:20px">The minimal inhibitory concentrations (MIC) of several important antimicrobial peptides GF-17, GF-18 and their |
| − | GF-17 was | + | variants were shown in Table 1. GF-17 was very efficient in eliminating both Gram-positive and Gram-negative |
| − | + | bacteria, such as S. aureus USA300and | |
| − | + | <i> E. coli</i> K-12in vitro (Wang et al., 2011). Additionally, to compare with LL-37, GF-17 is highly efficient | |
| − | + | in inhibition of biofilm formation and killing efficiency on | |
| + | <i>Staphylococcus aureus</i> (Fig 3c, table) ( Mishra et al., 2016; Wang et al., 2011 ).</p> | ||
</div> | </div> | ||
<div class="fig" align="center"> | <div class="fig" align="center"> | ||
<figure> | <figure> | ||
| − | <img width=" | + | <img width="650px" src="https://static.igem.org/mediawiki/2017/b/bc/Peptide_Production_table.png"> |
<figcaption> | <figcaption> | ||
| − | <strong>Table: Antimicrobial activity (MIC in μM) of GF-17, GE-18 and their variants (Wang et al., 2011)</figcaption> | + | <strong>Table 1: Antimicrobial activity (MIC in μM) of GF-17, GE-18 and their variants (Wang et al., 2011)</figcaption> |
</strong> | </strong> | ||
</figure> | </figure> | ||
<figure> | <figure> | ||
| − | <img width=" | + | <img width="650px" src="https://static.igem.org/mediawiki/2017/d/d5/Peptide_Production_3c.png"> |
<figcaption> | <figcaption> | ||
| − | <strong>Figure 3c: | + | <strong>Figure 3c: Anti-Staphylococcus aureus biofilm assay. Two different Staphylococcus aureus strains used: |
| − | and USA300. The result shows | + | USA200 and USA300. The result shows that GF-17 is more efficient in inhibiting the biofilm formation |
| − | + | of Staphylococcus aureus. | |
| + | </strong> | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| Line 142: | Line 238: | ||
<div class="para"> | <div class="para"> | ||
| − | <h1>GF-17 Reverse</h1> | + | <h1>GF-17 Reverse and GF-17 reverse peptide dimer</h1> |
| − | <p style="font-size:20px">GF-17 reverse | + | <p style="font-size:20px">GF-17 reverse (VLNRLFDKIRQVIRKFG) is a peptide which has a reverse peptide sequence of GF-17, and the GF-17 reverse |
| + | peptideadopts an alpha helical structure. The structure of GF-17 reverse peptide dimer (VLNRLFDKIRQVIRKFG-VLNRLFDKIRQVIRKFG) | ||
| + | is also α helical (Figure 4); (predicted by http://www.compbio.dundee.ac.uk/jpred/index.html). (Mishra et | ||
| + | al., 2016)). | ||
| + | </p> | ||
</div> | </div> | ||
| Line 149: | Line 249: | ||
<figure> | <figure> | ||
<img width="700px" src="https://static.igem.org/mediawiki/2017/e/ed/Peptide_Production_4.png"> | <img width="700px" src="https://static.igem.org/mediawiki/2017/e/ed/Peptide_Production_4.png"> | ||
| − | <figcaption>Figure 4: | + | <figcaption> |
| + | <strong>Figure 4: The secondary structure of GF-17 reverse dimer sequence, it seems that the dimer is α-helix. | ||
(predicted by http://www.compbio.dundee.ac.uk/jpred/index.html ) | (predicted by http://www.compbio.dundee.ac.uk/jpred/index.html ) | ||
| − | + | </strong> | |
| + | </figcaption> | ||
</figure> | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <h1>Grammistin-Pp1</h1> | ||
| + | <p style="font-size:20px">Grammistins are a group of peptides secreted by soapfishes and characterized by its amphiphilic -helical structure. | ||
| + | There are four types of Grammistins: Pp1,Pp3, Gs1, and Gs2. Gs 1 and Gs 2 are from | ||
| + | <i>Grammistes sexlineatus</i> and Pp 1 and Pp 3 from | ||
| + | <i>Pogonoperca punctata </i> (Yokota, Nagashima, and Shiom, 2001). Grammistin-Pp1 is made up of thirteen amino | ||
| + | acid residues. Studies showed that Grammistin-Pp1 could interact with phospholipids and influence the permeability | ||
| + | of membrane. Sugiyama et al. (2006) reported that the peptide Grammistin-Pp1 is effective on both Gram negative | ||
| + | bacteria and Gram positive bacteria, which includes </i>Staphylococcus aureus. </i> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <div class="fig" align="center"> | ||
| + | <figure> | ||
| + | <img width="650px" src="https://static.igem.org/mediawiki/2017/d/d8/Peptide_Production_5.png"> | ||
| + | <figcaption> | ||
| + | <strong>Figure 5: 2D structure of Grammistin-Pp1 (PubChem)</strong> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <h1>Experimental design on the peptide genes</h1> | ||
| + | <h2> | ||
| + | <strong>Using tandem repeats strategy</strong> | ||
| + | </h2> | ||
| + | <h2>Why using three copies in the tandem repeats?</h2> | ||
| + | <p style="font-size:20px">Our design (BBa_K2309028)using the tandem repeats strategy to express several copies of each anti-microbial peptide | ||
| + | (LL-37, GF-17, and Grammistin-Pp1). Two gene will recombinant together in tandem repeats. Replication slippage | ||
| + | could be formed between homology sequence between long space (50-100bp in general). | ||
| + | </p> | ||
| + | <p style="font-size:20px">In our project, we put repeat sequences together as ‘---AAA---BBB---CCC---‘ rather than ‘---ABC---ABC---ABC---‘ | ||
| + | strategy. Therefore, recombination could happened between homology sequence. The replication slippage will | ||
| + | not formed (Gemayel. et al, 2010).</p> | ||
| + | <h2>Why adding histidine-tag on LL-37?</h2> | ||
| + | <p style="font-size:20px">We added a 6x His tag at the end of LL-37 and cloned the three copies of the tagged LL-37 gene at the end of | ||
| + | the whole construct as shown in Figure 5 below. This design is used for the detection of the expression of | ||
| + | the peptides from the whole construct. It is very likely that if the peptide LL-37 plus the histidine tag | ||
| + | was detected by immunoblotting, that the other peptides located upstream of LL-37 (GF-17, Grammistin-Pp1, | ||
| + | and LL-37) were also translated .</p> | ||
| + | </div> | ||
| + | |||
| + | <div class="fig" align="center"> | ||
| + | <figure> | ||
| + | <img width="700px" src="https://static.igem.org/mediawiki/2017/b/bc/Peptide_Production_6.png"> | ||
| + | <figcaption> | ||
| + | <strong>Figure 6: the tandem repeats strategy.</strong> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <h2> | ||
| + | <strong>LL-37 + 6xHis tag (BBa_K2309022)</strong> | ||
| + | </h2> | ||
| + | <p style="font-size:20px">The purpose of coupling 6xHis-tag with LL-37can also be used for purifying the anti-microbial peptide produced | ||
| + | in the bacterial cells.6xHis tag is six histidine amino acids tandem arranged together which has a strong | ||
| + | affinity with the nickel ion in the Ni-NTA purification system. After purification, immunodot blot method | ||
| + | was used to confirm that LL-37 was produced.</p> | ||
| + | </div> | ||
| + | <div class="para"> | ||
| + | <h1>Final construction</h1> | ||
| + | </div> | ||
| + | |||
| + | <div class="fig" align="center"> | ||
| + | <figure> | ||
| + | <img width="700px" src="https://static.igem.org/mediawiki/2017/b/b4/Peptide_Production_8.png"> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <h1>Brief experimental protocols</h1> | ||
| + | <ol> | ||
| + | <li>Tests on the antimicrobial activities of the chemically synthesized peptides (LL-37, GF-17, and Grammistin-Pp1) | ||
| + | by: | ||
| + | <dl> | ||
| + | <dt>a. Inhibition ring assay</dt> | ||
| + | <dt>b. Minimal inhibitory concentration assay</dt> | ||
| + | <dt>c. Biofilm formation assay</dt> | ||
| + | <dt>d. Growth curve measurement (Spectrophotometric assay)</dt> | ||
| + | </dl> | ||
| + | </li> | ||
| + | <li>Construct the whole circuit to produce peptides</li> | ||
| + | <li>Using the dot blot assay and 6x His tag to purify and evaluate on the peptides produced by the whole circuit.</li> | ||
| + | </ol> | ||
| + | </div> | ||
| + | |||
| + | <div class="para"> | ||
| + | <h1>References</h1> | ||
| + | <ul> | ||
| + | <li>Wang. G., et al. (2011) ‘Decoding the Functional Roles of Cationic Side Chains of the Major Antimicrobial | ||
| + | Region of Human Cathelicidin LL-37’, | ||
| + | <i> Antimicrobial Agents and Chemotherapy</i>, pp. 845-856, ASM [Online]. DOI: 10.1128/AAC.05637-11 (Accessed: | ||
| + | 2017 August 28 | ||
| + | <sup>th</sup>)</li> | ||
| + | <li>Samperio.P (2010) ‘The human cathelicidin hCAP18/LL-37: A multifunctional peptide involved in mycobacterial | ||
| + | infections’, | ||
| + | <i>Peptides </i>, 31 (2010), pp. 1791-1798, ScienceDirect [Online]. DOI: 10.1016/j.peptides.2010.06.016 | ||
| + | (Accessed: 2017 August 28 | ||
| + | <sup>th</sup> )</li> | ||
| + | <li>Ramos. R., Domingues. L., Gama. M., (2011) ‘LL37, a human antimicrobial peptide with immunomodulatory properties’, | ||
| + | <i> | ||
| + | Science against microbial pathogens: communicating current research and technological advances</i>, | ||
| + | pp. 915-925, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 | ||
| + | <sup>th</sup>)</li> | ||
| + | <li>Dathe. M., Wieprecht. T., (1999) ‘Review Structural features of helical antimicrobial peptides: their potential | ||
| + | to modulate activity on model membranes and biological cells’, | ||
| + | <i> Biochimica et Biophysica Acta</i>, 1462 (1999), pp. 71-87, ScienceDirect [Online]. Available from: www.sciencedirect.com | ||
| + | (Accessed: 2017 August 24 | ||
| + | <sup>th</sup>)</li> | ||
| + | <li>Rocca. P., et al. (1999) ‘Review Simulation studies of the interaction of antimicrobial peptides and lipid | ||
| + | bilayers ’, | ||
| + | <i>Biochimica et Biophysica Acta</i>, 1462 (1999), pp. 185-200, ScienceDirect [Online]. Available from: | ||
| + | www.sciencedirect.com (Accessed: 2017 August 24 | ||
| + | <sup>th</sup>)</li> | ||
| + | <li> | ||
| + | Dürr. U., Sudheendra. U., Ramamoorthy. A., (2006) ‘Review LL-37, the only human member of the cathelicidin family of antimicrobial | ||
| + | peptides’, | ||
| + | <i>Biochimica et Biophysica Acta</i>, 1758 (2006), pp. 1408–1425, ScienceDirect [Online]. DOI: 10.1016/j.bbamem.2006.03.030 | ||
| + | (Accessed: 2017 August 24 | ||
| + | <sup>th</sup>) | ||
| + | </li> | ||
| + | <li> | ||
| + | Yokota, H., Nagashima, Y., and Shiom, K., (2001) ‘Interaction of grammistins with lipids and their antibacterial activity’, | ||
| + | <i> | ||
| + | Fisheries Science</i>, 2001 (67), pp. 928-933, ScienceDirect [Online]. Available from: http://www.sciencedirect.com | ||
| + | (Accessed: 2017 August 26 | ||
| + | <sup>th</sup>) | ||
| + | </li> | ||
| + | <li> | ||
| + | Sugiyama, N. et al. (2006) ‘Biological activities of synthetic grammistins and analogous peptides’, | ||
| + | <i>Fisheries Science</i>, 2006 (72), pp. 1114-1120, ScienceDirect [Online]. Available from: http://www.sciencedirect.com(Accessed: | ||
| + | 2017 August 26 | ||
| + | <sup>th</sup>) | ||
| + | </li> | ||
| + | <li> | ||
| + | Bahar, A., Ren, D., (2013) ‘Antimicrobial Peptides’, | ||
| + | <i>pharmaceuticals, </i> 2013 (6), pp. 1543-1575, NCBI [Online]. DOI: 10.3390/ph6121543. | ||
| + | </li> | ||
| + | <li> | ||
| + | Hancock, R & Sahl, H., (2006) ‘Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies’, | ||
| + | <i>Nature biotechnology, </i> 24 (12), Nature [Online]. DOI: :10.1038/nbt1267 | ||
| + | </li> | ||
| + | <li> | ||
| + | Gemayel, R., (2010) ‘Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences’, | ||
| + | <i>Annual Review of Genetics, </i> ResearchGate [Online]. DOI: 10.1146/annurev-genet-072610-155046 | ||
| + | </li> | ||
| + | </ul> | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | <!-- banner --> | ||
| + | <div class="text-center" style="background:#cfe5d9;"> | ||
| + | <h1 style="font-weight:bolder;color:#006934; ">Collaborators and Supporters</h1> | ||
| + | <div class="container-fluid supporters-logos"> | ||
| + | <div class="row" style="padding-bottom:10px"> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.synbio-tech.com.cn"> | ||
| + | <img class="" src="https://static.igem.org/mediawiki/2017/3/38/Synbio_tech_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.wx2h.com/web/index.php"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/f/f7/Wuxi_no2_hospital_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.chinapeptides.qianyan.biz"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/7/7d/Qiangyao_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="https://www.neb.com"> | ||
| + | <img class="" src="https://static.igem.org/mediawiki/2017/0/06/NEB_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="https://www.snapgene.com"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/c/cb/Snapgene_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.genscript.com"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/9/9b/Genscript.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!-- /.container --!> | ||
| + | </div><!-- /.text-center --!> | ||
| + | |||
| + | <!-- footer --> | ||
| + | <footer> | ||
| + | <div class="text-center"> | ||
| + | <div class="container-fluid"> | ||
| + | <div class="row"> | ||
| + | <div class="col-md-4 loaction"> | ||
| + | <h4>Location</h4> | ||
| + | <p style="text-align:center;">Rm 363, Science Building | ||
| + | <br> Xi'an Jiaotong-Liverpool University | ||
| + | <br> 111 Ren'ai Road, Suzhou, China | ||
| + | <br> 215123 | ||
| + | </p> | ||
| + | </div> | ||
| + | <div class="col-md-4 social"> | ||
| + | <h4>Social</h4> | ||
| + | <a href=""> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/9/9f/XJTLU_facebook.png" alt="facebook" width=30 height=30> | ||
| + | </a> | ||
| + | <a href=""> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/7/72/XJTLU_blog.png" alt="blog" width=30 height=30> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="col-md-4 contact"> | ||
| + | <h4>Get in touch</h4> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/1/19/XJTLU_email.png" alt="emali" width=30 height=30> | ||
| + | <p style="text-align:center;">igem@xjtlu.edu.cn</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="text-center" style="background:#003b73;"> | ||
| + | <p style="text-align:center; color:white;">XJTLU-CHINA iGEM 2017</p> | ||
| + | </div> | ||
| + | </footer> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 04:13, 27 November 2017
Peptide Production
The peptide production contains two parts: cross-inhibition and antimicrobial peptides
Cross-inhibition
As introduced in the description section, there are four different classes in the Agr quorum sensing systems which are referred to as Agr-I, Agr-II, Agr-III, and Agr-IV and each is capable of recognizing a unique AIP structure, which is AIP-I, AIP-II, AIP-III and AIP-IV, respectively. S. aureus may also be classified into four groups/strains (I to IV) according to the class of the AIPs produced. Similarly, the proteins involved in signal biosynthesis (AgrB and AgrD) and surface receptor binding (AgrC) also show variability. Interestingly, different AIP signals cross-inhibit the activity of the others.

For the cross-inhibition, we introduce two genes, agrB and agrD from a different type of S.aureus. The transmembrane protein AgrB cleaves the precursor peptide AgrB into a different type of AIPs which is capable of inhibiting the combination of the original AIPs with the membrane AgrC, and thus stopping the quorum sensing and growth of biofilms. The construction is shown below.

Antimicrobial Peptide

Anti-microbial peptide (AMP) is a part of the innate immune system of most multi-cellular organisms to counter microbial infections (Margitta and Torsten, 1999). Among these structural groups, α-helix and β-sheet structures are more common. AMPs kill cells by disrupting membrane integrity (via interaction with the negatively charged molecules on the cell membrane), by inhibiting proteins, DNA and RNA synthesis, or by interacting with certain intracellular targets. All AMPs known by the late-90s are cationic. However, the concept that AMPs need to be cationic was changed later with the discovery of negatively charged AMPs in 1997. For example maximin-H5 from frog skin and dermicidin secreted from the sweat gland tissues of humans are both anionic peptides (Bahar, and Ren, 2013).
One of important features of AMPs is their rapid killing effect. AMPs can influence cell membrane in few seconds after contact. AMPs are also known to enhance the activities of antibiotics through their synergistic effects. For example, the combination of penicillin with pediocin and ampicillin with nisin Z exhibited a killing effect on Pseudomonas fluorescens with 13- and 155-fold lower in the minimum inhibitory concentration (MIC), respectively, compared to using the antibiotics alone (Bahar, and Ren, 2013).
Another feature of AMPs is their activation on the immune response defense. As shown by Figure 1c, the role of cationic host-defense peptides in modulating the innate immune response and boosting infection-resolving immunity while dampening potentially harmful pro-inflammatory responses gives these peptides the potential to become an entirely new therapeutic approach against bacterial infections(Hancock & Sahl, 2006).

Although bacteria have diverse mechanisms of resistance to AMPs, it is encouraging to notice that the general lipid bilayer structure of bacterial membranes makes it hard to develop a complete resistance against AMPs. Also, the resistance against AMPs reported to date is not as strong as those against antibiotics and it is only found in a limited number of AMPs.
Cationic and amphipathic antimicrobial Peptides
The cationic and amphipathic α-helical structure is the most common conformation in these types of peptides but some hydrophobic α-helical peptides can also possess antimicrobial activity. In this project we choose three different cationic antimicrobial peptides with α-helical conformation.
Figure 1d shows the molecular mechanism of cationic AMPs containing α-helical structure. Most of the cationic AMPs associate with the charged lipid group of the bacteria plasma membrane. The α-helical structure disrupts the packing of lipid molecules such that the membrane becomes leaky (Rocca et al.,1999).

LL-37
LL-37 is the only cathelicidin-derived antimicrobial peptide found in humans (Dürr, Sudheendra and Ramamoorthy, 2006). Mature LL-37 has 37 amino acid residues starting with two leucines (NH2-LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES-COOH). The peptide is cleaved from a larger protein, hCAP-18 by extracellular proteolysis of proteinase 3 from the C-terminal end of hCAP18 (Patricia, 2010; Ramos, Domingues, and Gama, 2011). The peptide composed of two mainly parts: from residue Leu2 to Leu31, which is a α-helical structure (Fig 2b) and a 6 residues form the loop structure at the terminus(Fig 2a).
Ramos, Domingues, and Gama (2011) also reported that LL-37 has additional roles such as regulating the inflammatory response in wounds or infection sites, binding and neutralizing LPS, and wound closure, apart from anti-microbial property (Figure 2c).



GF-17
GF-17 is a highly efficient anti-microbial peptide derived from the residure Phe-17 to Val-32 of LL-37(Fig 3a).


The minimal inhibitory concentrations (MIC) of several important antimicrobial peptides GF-17, GF-18 and their variants were shown in Table 1. GF-17 was very efficient in eliminating both Gram-positive and Gram-negative bacteria, such as S. aureus USA300and E. coli K-12in vitro (Wang et al., 2011). Additionally, to compare with LL-37, GF-17 is highly efficient in inhibition of biofilm formation and killing efficiency on Staphylococcus aureus (Fig 3c, table) ( Mishra et al., 2016; Wang et al., 2011 ).


GF-17 Reverse and GF-17 reverse peptide dimer
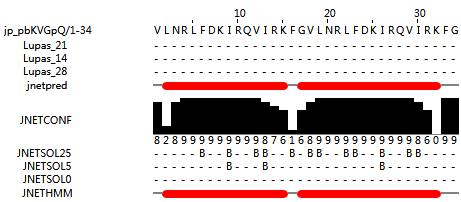
GF-17 reverse (VLNRLFDKIRQVIRKFG) is a peptide which has a reverse peptide sequence of GF-17, and the GF-17 reverse peptideadopts an alpha helical structure. The structure of GF-17 reverse peptide dimer (VLNRLFDKIRQVIRKFG-VLNRLFDKIRQVIRKFG) is also α helical (Figure 4); (predicted by http://www.compbio.dundee.ac.uk/jpred/index.html). (Mishra et al., 2016)).
Grammistin-Pp1
Grammistins are a group of peptides secreted by soapfishes and characterized by its amphiphilic -helical structure. There are four types of Grammistins: Pp1,Pp3, Gs1, and Gs2. Gs 1 and Gs 2 are from Grammistes sexlineatus and Pp 1 and Pp 3 from Pogonoperca punctata (Yokota, Nagashima, and Shiom, 2001). Grammistin-Pp1 is made up of thirteen amino acid residues. Studies showed that Grammistin-Pp1 could interact with phospholipids and influence the permeability of membrane. Sugiyama et al. (2006) reported that the peptide Grammistin-Pp1 is effective on both Gram negative bacteria and Gram positive bacteria, which includes Staphylococcus aureus.

Experimental design on the peptide genes
Using tandem repeats strategy
Why using three copies in the tandem repeats?
Our design (BBa_K2309028)using the tandem repeats strategy to express several copies of each anti-microbial peptide (LL-37, GF-17, and Grammistin-Pp1). Two gene will recombinant together in tandem repeats. Replication slippage could be formed between homology sequence between long space (50-100bp in general).
In our project, we put repeat sequences together as ‘---AAA---BBB---CCC---‘ rather than ‘---ABC---ABC---ABC---‘ strategy. Therefore, recombination could happened between homology sequence. The replication slippage will not formed (Gemayel. et al, 2010).
Why adding histidine-tag on LL-37?
We added a 6x His tag at the end of LL-37 and cloned the three copies of the tagged LL-37 gene at the end of the whole construct as shown in Figure 5 below. This design is used for the detection of the expression of the peptides from the whole construct. It is very likely that if the peptide LL-37 plus the histidine tag was detected by immunoblotting, that the other peptides located upstream of LL-37 (GF-17, Grammistin-Pp1, and LL-37) were also translated .

LL-37 + 6xHis tag (BBa_K2309022)
The purpose of coupling 6xHis-tag with LL-37can also be used for purifying the anti-microbial peptide produced in the bacterial cells.6xHis tag is six histidine amino acids tandem arranged together which has a strong affinity with the nickel ion in the Ni-NTA purification system. After purification, immunodot blot method was used to confirm that LL-37 was produced.
Final construction

Brief experimental protocols
- Tests on the antimicrobial activities of the chemically synthesized peptides (LL-37, GF-17, and Grammistin-Pp1)
by:
- a. Inhibition ring assay
- b. Minimal inhibitory concentration assay
- c. Biofilm formation assay
- d. Growth curve measurement (Spectrophotometric assay)
- Construct the whole circuit to produce peptides
- Using the dot blot assay and 6x His tag to purify and evaluate on the peptides produced by the whole circuit.
References
- Wang. G., et al. (2011) ‘Decoding the Functional Roles of Cationic Side Chains of the Major Antimicrobial Region of Human Cathelicidin LL-37’, Antimicrobial Agents and Chemotherapy, pp. 845-856, ASM [Online]. DOI: 10.1128/AAC.05637-11 (Accessed: 2017 August 28 th)
- Samperio.P (2010) ‘The human cathelicidin hCAP18/LL-37: A multifunctional peptide involved in mycobacterial infections’, Peptides , 31 (2010), pp. 1791-1798, ScienceDirect [Online]. DOI: 10.1016/j.peptides.2010.06.016 (Accessed: 2017 August 28 th )
- Ramos. R., Domingues. L., Gama. M., (2011) ‘LL37, a human antimicrobial peptide with immunomodulatory properties’, Science against microbial pathogens: communicating current research and technological advances, pp. 915-925, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 th)
- Dathe. M., Wieprecht. T., (1999) ‘Review Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells’, Biochimica et Biophysica Acta, 1462 (1999), pp. 71-87, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 th)
- Rocca. P., et al. (1999) ‘Review Simulation studies of the interaction of antimicrobial peptides and lipid bilayers ’, Biochimica et Biophysica Acta, 1462 (1999), pp. 185-200, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 th)
- Dürr. U., Sudheendra. U., Ramamoorthy. A., (2006) ‘Review LL-37, the only human member of the cathelicidin family of antimicrobial peptides’, Biochimica et Biophysica Acta, 1758 (2006), pp. 1408–1425, ScienceDirect [Online]. DOI: 10.1016/j.bbamem.2006.03.030 (Accessed: 2017 August 24 th)
- Yokota, H., Nagashima, Y., and Shiom, K., (2001) ‘Interaction of grammistins with lipids and their antibacterial activity’, Fisheries Science, 2001 (67), pp. 928-933, ScienceDirect [Online]. Available from: http://www.sciencedirect.com (Accessed: 2017 August 26 th)
- Sugiyama, N. et al. (2006) ‘Biological activities of synthetic grammistins and analogous peptides’, Fisheries Science, 2006 (72), pp. 1114-1120, ScienceDirect [Online]. Available from: http://www.sciencedirect.com(Accessed: 2017 August 26 th)
- Bahar, A., Ren, D., (2013) ‘Antimicrobial Peptides’, pharmaceuticals, 2013 (6), pp. 1543-1575, NCBI [Online]. DOI: 10.3390/ph6121543.
- Hancock, R & Sahl, H., (2006) ‘Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies’, Nature biotechnology, 24 (12), Nature [Online]. DOI: :10.1038/nbt1267
- Gemayel, R., (2010) ‘Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences’, Annual Review of Genetics, ResearchGate [Online]. DOI: 10.1146/annurev-genet-072610-155046







