| Line 293: | Line 293: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | <!-- banner --> | ||
| + | <div class="text-center" style="background:#cfe5d9;"> | ||
| + | <h1 style="font-weight:bolder;color:#006934; ">Collaborators and Supporters</h1> | ||
| + | <div class="container-fluid supporters-logos"> | ||
| + | <div class="row" style="padding-bottom:10px"> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.synbio-tech.com.cn"> | ||
| + | <img class="" src="https://static.igem.org/mediawiki/2017/3/38/Synbio_tech_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.wx2h.com/web/index.php"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/f/f7/Wuxi_no2_hospital_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.chinapeptides.qianyan.biz"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/7/7d/Qiangyao_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="https://www.neb.com"> | ||
| + | <img class="" src="https://static.igem.org/mediawiki/2017/0/06/NEB_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="https://www.snapgene.com"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/c/cb/Snapgene_logo.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-md-4 col-sm-6"> | ||
| + | <a href="http://www.genscript.com"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/9/9b/Genscript.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!-- /.container --!> | ||
| + | </div><!-- /.text-center --!> | ||
| + | |||
| + | <!-- footer --> | ||
| + | <footer> | ||
| + | <div class="text-center"> | ||
| + | <div class="container-fluid"> | ||
| + | <div class="row"> | ||
| + | <div class="col-md-4 loaction"> | ||
| + | <h4>Location</h4> | ||
| + | <p style="text-align:center;">Rm 363, Science Building | ||
| + | <br> Xi'an Jiaotong-Liverpool University | ||
| + | <br> 111 Ren'ai Road, Suzhou, China | ||
| + | <br> 215123 | ||
| + | </p> | ||
| + | </div> | ||
| + | <div class="col-md-4 social"> | ||
| + | <h4>Social</h4> | ||
| + | <a href=""> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/9/9f/XJTLU_facebook.png" alt="facebook" width=30 height=30> | ||
| + | </a> | ||
| + | <a href=""> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/7/72/XJTLU_blog.png" alt="blog" width=30 height=30> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="col-md-4 contact"> | ||
| + | <h4>Get in touch</h4> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/1/19/XJTLU_email.png" alt="emali" width=30 height=30> | ||
| + | <p style="text-align:center;">igem@xjtlu.edu.cn</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="text-center" style="background:#003b73;"> | ||
| + | <p style="text-align:center; color:white;">XJTLU-CHINA iGEM 2017</p> | ||
| + | </div> | ||
| + | </footer> | ||
</body> | </body> | ||
</html> | </html> | ||
Revision as of 09:57, 30 October 2017
Peptide Production
Anti-Microbial Peptide
Anti-microbial peptide (AMP) is a part of the innate immune system of most multi-cellular organisms to counter microbial infections (Margitta and Torsten, 1999). The cationic and amphipathic α-helix structure is the most wildly conformation in those peptides but some hydrophobic α-helical peptides which possess antimicrobial activity. This year we choose three different cationic antimicrobial peptides which encompass α-helical conformation in our project.
Figure 1 shows the molecular mechanism of cationic AMPs α-helical structure. Most of cationic AMPs associate with lipid group of bacteria membrane. The α-helical structure disrupt the packing of lipid molecules such that the membrane becomes leaky (Rocca et al., 1999).

LL-37
LL-37 is the only cathelicidin-derived antimicrobial peptide found in humans (Dürr, Sudheendra and Ramamoorthy, 2006). Mature LL-37 has 37 amino acid residues starting with two leucines (NH2-LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES-COOH). The peptide is cleaved from a larger protein, hCAP-18 by extracellular proteolysis of proteinase 3 from the C-terminal end of hCAP18 (Patricia, 2010; Ramos, Domingues, and Gama, 2011). The peptide composed of two mainly parts: from residue Leu2 to Leu31 is α-helical structure (Fig 2b) and 6 residues form loop structure (Fig 2a).
Ramos, Domingues, and Gama (2011) also reported that LL-37 has additional roles such as regulating the inflammatory response to wound or infection sites, binding and neutralizing LPS, and wound closure apart from anti-microbial property (Figure 2c).



GF-17
GF-17 is a high efficiency anti-microbial peptide which modified from LL-37 residue Phe-17 to Val-32 (Fig 3a).


Table shows the Minimal Inhibitory concentration (MIC) of Anti-microbial peptides GF-17, GF-18 and their variants. GF-17 was capable of eliminating both Gram-positive and Gram-negative bacteria, such as S. aureus USA300 and E. coli K-12 in vitro (Wang et al., 2011). Additionally, to compare with LL-37, GF-17 has highly efficiency on anti-Staphylococcus aureus biofilm and killing efficiency (Fig 3c, table) ( Mishra et al., 2016; Wang et al., 2011 ).


GF-17 Reverse
GF-17 reverse sequence is reverse from GF-17, the structure of GF-17 reverse sequence dimer is semi-α helix.
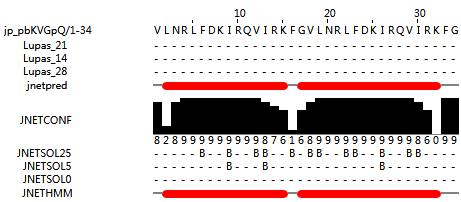
Grammistin-Pp1
Grammistin-Pp1 is an AMP found in skin secretions from the clown grouper fish and is made up of thirteen amino acid residues. This AMP is used primarily to fend off predators. Although initially thought to have little antimicrobial properties, studies proved that grammistin-Pp1 target cell membranes by interacting with phospholipids. Grammistin-Pp1 has been shown to be lethal against many gram-negative and gram-positive bacteria including S. aureus. A primary reason for our selection of Grammistin-Pp1 was the high concentration needed to inhibit the growth of E. coli. (Yokota et al., 2001)

Experiment design
Three copies of tandem repeats strategy (BBa_K2309028)
Why Three copies tandem repeats?
Our design using tandem repeats strategy to express several copies of anti-microbial peptide (LL-37, GF-17, and Grammistin-Pp1)
Why LL-37+6x His tag?
We add 6x His tag at the end of LL-37 and the position of LL-37 with 6x His tag is at the end of whole pathway as Figure 5 shows below. This design is used for evaluating the LL-37 is produced by whole pathway. Once peptide LL-37 plus his tag was detected, we assume that peptides before LL-37 (GF-17, Grammistin-Pp1, and LL-37) is produced.

LL-37 + 6xHis tag (BBa_K2309022)
The purpose of the LL-37 with 6x His-tag is used for purifying the anti-microbial peptide production. 6xHis tag is six histidine amino acids tandem together which has strong affinity with Ni-NTA purification system. After purification, running SDS-PAGE and using dot blot method to confirm the LL-37 were produced.

Final construction

Experiment brief protocol
-
- a. Inhibition Ring
- b. Minimal Inhibitory concentration assay
- c. Biofilm assay
- d. Growth curve measurement
- Construct bio circuit to produce peptides
- Using dot blot and 6x His tag to purify and evaluate peptides can be produced by bio circuit.
Reference
- Wang. G., et al. (2011) ‘Decoding the Functional Roles of Cationic Side Chains of the Major Antimicrobial Region of Human Cathelicidin LL-37’, Antimicrobial Agents and Chemotherapy, pp. 845-856, ASM [Online]. DOI: 10.1128/AAC.05637-11 (Accessed: 2017 August 28 th)
- Samperio.P (2010) ‘The human cathelicidin hCAP18/LL-37: A multifunctional peptide involved in mycobacterial infections’, Peptides , 31 (2010), pp. 1791-1798, ScienceDirect [Online]. DOI: 10.1016/j.peptides.2010.06.016 (Accessed: 2017 August 28 th )
- Ramos. R., Domingues. L., Gama. M., (2011) ‘LL37, a human antimicrobial peptide with immunomodulatory properties’, Science against microbial pathogens: communicating current research and technological advances, pp. 915-925, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 th)
- Dathe. M., Wieprecht. T., (1999) ‘Review Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells’, Biochimica et Biophysica Acta, 1462 (1999), pp. 71-87, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 th)
- Rocca. P., et al. (1999) ‘Review Simulation studies of the interaction of antimicrobial peptides and lipid bilayers ’, Biochimica et Biophysica Acta, 1462 (1999), pp. 185-200, ScienceDirect [Online]. Available from: www.sciencedirect.com (Accessed: 2017 August 24 th)
- Dürr. U., Sudheendra. U., Ramamoorthy. A., (2006) ‘Review LL-37, the only human member of the cathelicidin family of antimicrobial peptides’, Biochimica et Biophysica Acta, 1758 (2006), pp. 1408–1425, ScienceDirect [Online]. DOI: 10.1016/j.bbamem.2006.03.030 (Accessed: 2017 August 24 th)







