| (11 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{Aix-Marseille|title=Detection of the disease|toc= | + | {{Aix-Marseille|title=Detection of the disease|toc=__noTOC__}} |
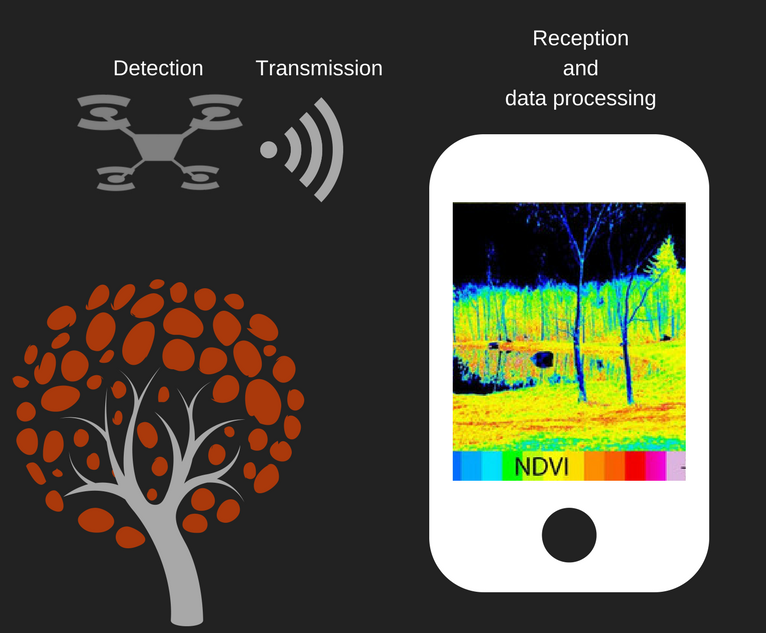
[[File:T--Aix-Marseille--transmission chain-1.png|right|400px|The hydric stress detection method.]] | [[File:T--Aix-Marseille--transmission chain-1.png|right|400px|The hydric stress detection method.]] | ||
| − | '''KILL XYL''' not only | + | [[Team:Aix-Marseille/Project|'''KILL XYL''']] aims not only to cure the disease caused by [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']], but also to detect it. Nowadays, the most effective way to detect the bacteria is a method called PCR (Polymerase Chain Reaction). |
| + | However, this method is complex and requires DNA samples from trees and lengthy and complex laboratory treatments. | ||
| − | Therefore, we | + | Therefore, we focused on another method which will allow us to work more easily in the open with hundreds of acres of crops. |
| + | For this, we thought about detecting the first symptoms of the disease: hydric stress. | ||
| + | The easiest way to measure the hydric stress is by assessing leaf dryness. | ||
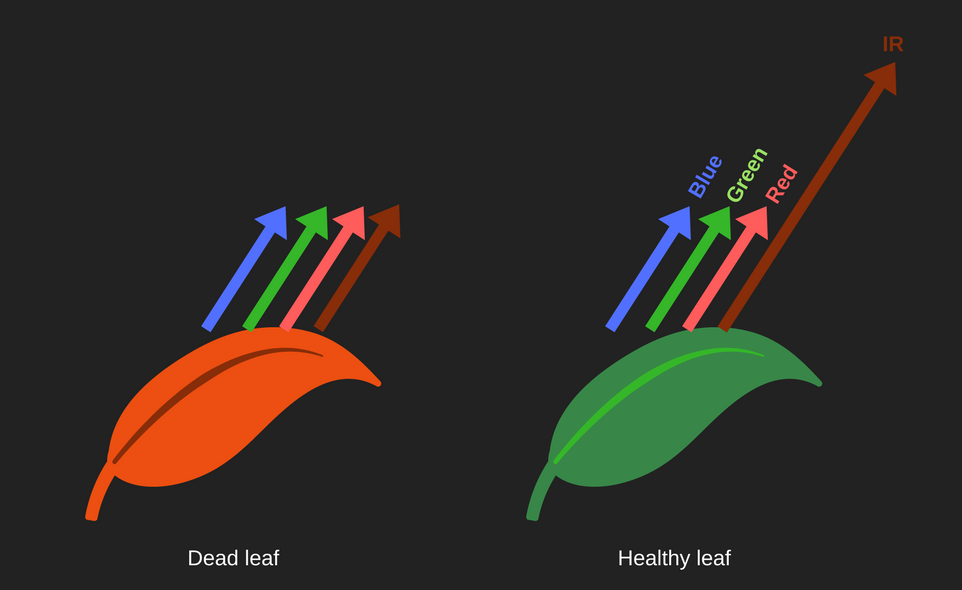
| − | [[File:T--Aix-Marseille--refraction spectrum leaves-1.png|400px|right]] | + | [[File:T--Aix-Marseille--refraction spectrum leaves-1.png|400px|right|thumb|Healthy leaves, because of photosynthesis, refract more infrared (IR) light than dry leaves.]] |
| − | Olive trees are evergreen which means that leaves will not dry naturally. However, dry leaves can be a consequence of other factors than the disease caused[[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']]. Thus our solution serves primarily as a warning device, it's appropriate to the detection of hydric stress | + | Olive trees are evergreen which means that leaves will not dry naturally. |
| + | However, dry leaves can be a consequence of other factors than the disease caused by [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']]. | ||
| + | Thus our solution serves primarily as a warning device, it's appropriate to the detection of hydric stress in crops. | ||
| + | If any is detected, then DNA samples should be taken to verify the presence of the bacteria. | ||
[[File:T--Aix-Marseille--NDVI comparison-1.jpg|400px|right|thumb|K-State Research and Extension | [[File:T--Aix-Marseille--NDVI comparison-1.jpg|400px|right|thumb|K-State Research and Extension | ||
Soybean NVDI photo]] | Soybean NVDI photo]] | ||
| − | + | As healthy leaves reflect more infrared light than dry leaves we can calculate the Normalized Difference Vegetation Index (NDVI), which allows determining the relative level of photosynthesis of a plant. NDVI is calculated by : $$\text{NDVI} = \frac{\text{NIR}-\text{RED}}{\text{NIR}+\text{RED}}$$ | |
| − | The calculated index is between -1 and 1 and is associated with a color scale (1 | + | The calculated index is between -1 and 1 and is associated with a color scale (1, red and -1, blue) that allows you to observe easily and quickly if the tree is healthy or not. |
==Detection== | ==Detection== | ||
| − | First, we | + | First, we built the camera. |
| + | For this, we used a Raspberry Pi 2 which supports the necessary acquisition and treatment software and a Black Pi camera. | ||
| + | The infrared blocking filter was removed from the camera and replaced with a Blue filter (ROSCO#2007). | ||
| + | This modification allows us to detect the NIR and RED light necessary to construct the NDVI image. | ||
==Software== | ==Software== | ||
| − | + | [[File:T--Aix-Marseille--NDVI-process.png|right|500px|thumb|]] | |
| − | + | We used Python and OpenCV to calculate our NDVI images. | |
| − | + | NDVI index computation is done pixel by pixel on the image to create a new grayscale image, which is then shown with a linear colormap (jet) for a better visual contrast. | |
| − | + | ||
Latest revision as of 02:29, 2 November 2017
Detection of the disease
KILL XYL aims not only to cure the disease caused by Xylella fastidiosa, but also to detect it. Nowadays, the most effective way to detect the bacteria is a method called PCR (Polymerase Chain Reaction). However, this method is complex and requires DNA samples from trees and lengthy and complex laboratory treatments.
Therefore, we focused on another method which will allow us to work more easily in the open with hundreds of acres of crops. For this, we thought about detecting the first symptoms of the disease: hydric stress. The easiest way to measure the hydric stress is by assessing leaf dryness.
Olive trees are evergreen which means that leaves will not dry naturally. However, dry leaves can be a consequence of other factors than the disease caused by X. fastidiosa. Thus our solution serves primarily as a warning device, it's appropriate to the detection of hydric stress in crops. If any is detected, then DNA samples should be taken to verify the presence of the bacteria.
As healthy leaves reflect more infrared light than dry leaves we can calculate the Normalized Difference Vegetation Index (NDVI), which allows determining the relative level of photosynthesis of a plant. NDVI is calculated by : $$\text{NDVI} = \frac{\text{NIR}-\text{RED}}{\text{NIR}+\text{RED}}$$
The calculated index is between -1 and 1 and is associated with a color scale (1, red and -1, blue) that allows you to observe easily and quickly if the tree is healthy or not.
Detection
First, we built the camera. For this, we used a Raspberry Pi 2 which supports the necessary acquisition and treatment software and a Black Pi camera. The infrared blocking filter was removed from the camera and replaced with a Blue filter (ROSCO#2007). This modification allows us to detect the NIR and RED light necessary to construct the NDVI image.
Software
We used Python and OpenCV to calculate our NDVI images. NDVI index computation is done pixel by pixel on the image to create a new grayscale image, which is then shown with a linear colormap (jet) for a better visual contrast.