|
|
| (45 intermediate revisions by 6 users not shown) |
| Line 1: |
Line 1: |
| | {{Aix-Marseille|title=Engineering bacteriophages|toc=__TOC__}} | | {{Aix-Marseille|title=Engineering bacteriophages|toc=__TOC__}} |
| | | | |
| − | ==M13 phages==
| + | [[File:T--Aix-Marseille--Phage-2.png|450px|right|thumb|Phage-like particles process.]] |
| | | | |
| − | M13 is a filamentous phage that infects E. coli that carry the F-episome. Active infection with M13 does not kill the host cell. The M13 phage particle consists of a single-stranded DNA (ssDNA) genome encased in approximately 2700 copies
| |
| − | of a major coat protein protein VIII.<ref name=Smeal>Smeal, S. W., Schmitt, M. A., Pereira, R. R., Prasad, A. & Fisk, J. D. Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation. Virology 500, 259–274 (2017).</ref>
| |
| | | | |
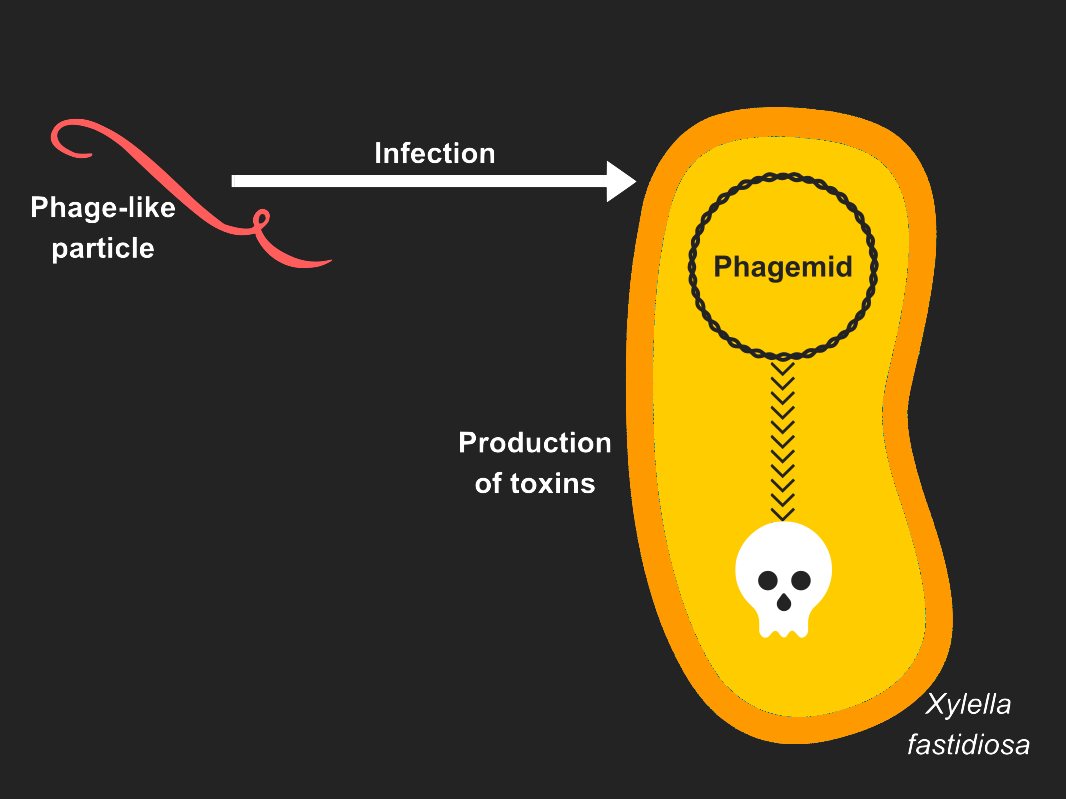
| − | ===Life cycle=== | + | Bacteriophages play a special role in the development of nanoscale cargo-delivery because they can be regarded as naturally occurring nanomaterials. |
| | + | We chose to explore Phage-like particles (PLPs)<ref name="Roldão">Roldão, A., Silva, A. C., Mellado, M. C. M., Alves, P. M. & Carrondo, M. J. T. Viruses and Virus-Like Particles in Biotechnology: Fundamentals and Applications. in Reference Module in Life Sciences (Elsevier, 2017).</ref> for use against pathogenic bacteria like [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']]. |
| | + | Our goal is to use a phage-like [[Team:Aix-Marseille/M13|M13]] to inject a lethal toxin into the bacterium. |
| | + | To help our research we asked [[Team:Aix-Marseille/HP/Interviews|Mireille Ansaldi]], a local phage expert. |
| | | | |
| − | [[File:T--Aix-Marseille--M13LCb.png|300px|right|thumb|M13 phage life cycle in ''Escherichia coli''.]] | + | Phage systems, like [[Team:Aix-Marseille/M13|M13]], have been widely employed in biotechnology, |
| | + | most prominently in the identification and selection of high affinity molecules through phage display <ref>Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).</ref>. |
| | + | The use of phages in materials science and nanotechnology is mainly due to their nanoscale size and simple life cycles. |
| | + | We decided to profit from this wide application in order to target [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] <ref name="Buttimer">Buttimer, C. et al. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 8, (2017).</ref>. |
| | | | |
| − | The M13 life cycle begins with passage of the phage genome into a host cell in a process induced by protein III (pIII). First, the single-strand DNA (ssDNA) is converted in double-strand DNA (dsDNA) by pII and pX, which allow the production of phage's protein.
| + | ==Phage-like particle design== |
| | | | |
| − | After a while, as the concentrations of phage proteins increase and the ssDNA is back converted as dsDNA. The protein V (pV) binds to the ssDNA genomes for packaging into progeny phages. It recognise the single stranded M13 origin of replication. The pV-sequestered ssDNA is recognized by the membrane spanning phage assembly complex. <ref name=Smeal>Smeal, S. W., Schmitt, M. A., Pereira, R. R., Prasad, A. & Fisk, J. D. Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation. Virology 500, 259–274 (2017).</ref>
| + | Bacteriophages are able to generate new copies of themselves using the information in their genome <ref>Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).</ref>. |
| | + | In our design, we decided to limit the phage's ability to reproduce in order to contain it, as our interview with [[Team:Aix-Marseille/HP/Interviews|Jacques VAN HELDEN]] indicated this was an important point. |
| | + | There are already well-tried methods to reduce the reproductive capacity of phages as well as to make phages with foreign proteins<ref name= "Roldão" />. |
| | | | |
| − | ===Protein III=== | + | Phage-like particles (PLPs) mimic the organization of native phages but lack the viral genome. |
| | + | They have been used as vehicles for drug and gene delivery and also as tools in nanobiotechnology <ref name="Czapar">Czapar, A. E. & Steinmetz, N. F. Plant viruses and bacteriophages for drug delivery in medicine and biotechnology. Current Opinion in Chemical Biology 38, 108–116 (2017). </ref>. |
| | + | In this project, we create [[Team:Aix-Marseille/M13|M13]] phage-like particles that inject toxin genes into target bacteria. |
| | | | |
| − | [[File:T--Aix-Marseille--M13p3b.png|400px|right|thumb|Description of the domains composing M13's protein III.]]
| + | ==Phage-like particle in the environment== |
| | | | |
| − | The molecular interactions that mediate the entry of ''Escherichia coli'' derived filamentous phages into their hosts have been studied in considerable detail. The 424-amino-acid protein III is thought to consist of a leader sequence and three domains, separated by glycine-rich regions, that serve distinct roles in phage entry and release. The first two pIII domains, D1 and D2, are required for M13 adsorption and entry, while the third domain D3 is required for the assembly and release of M13 particles from host.<ref name="Heilpern">Heilpern, A. J. & Waldor, M. K. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185, 1037–1044 (2003).</ref>
| + | Beyond efficiency, we want our product to be environmentally friendly so that it doesn't contaminate nature with GMO's. |
| | + | To investigate environmental impact, we used soil samples to test the longevity of our phages in nature. |
| | + | We are also working with the [http://biam.cea.fr/drf/biam/Pages/laboratoires/lgbp.aspx LGBP lab], where we test our phage in the model organism ''Arabidopsis thaliana''. |
| | | | |
| − | ==Design==
| + | <div class="row-icons white-icons"> |
| − | | + | * [[File:T--Aix-Marseille--Pdesign.png|link=Team:Aix-Marseille/M13_Design]]<span class="legend">Phage design</span> |
| − | ===Phages-like particules===
| + | * [[File:T--Aix-Marseille--plant.png|link=Team:Aix-Marseille/M13_test]]<span class="legend">Environmental test</span> |
| − | | + | </div> |
| − | Bacteriophages are capable of expressing their genomes, and generating new copies of themselves. We choose to limit the phage ability to reproduce itself in order to contain it. As it is possible to produce recombinant viruses that express foreign proteins, it is possible to restrain their capacity to reproduce themself. <ref name= Silva>Roldão, A., Silva, A. C., Mellado, M. C. M., Alves, P. M. & Carrondo, M. J. T. Viruses and Virus-Like Particles in Biotechnology: Fundamentals and Applications. in Reference Module in Life Sciences (Elsevier, 2017).</ref>
| + | |
| − | | + | |
| − | Virus-like particles (VLPs) are multiprotein structures that mimic the organization and conformation of authentic native viruses but lack the viral genome. They have been applied not only as prophylactic and therapeutic vaccines but also as vehicles in drug and gene delivery and, more recently, as tools in nanobiotechnology. <ref name= Silva>Roldão, A., Silva, A. C., Mellado, M. C. M., Alves, P. M. & Carrondo, M. J. T. Viruses and Virus-Like Particles in Biotechnology: Fundamentals and Applications. in Reference Module in Life Sciences (Elsevier, 2017).</ref>
| + | |
| − | ===Engineering M13===
| + | |
| − | [[File:T--Aix-Marseille--M13K07.png|350px|right]] | + | |
| − | | + | |
| − | In order to engineered multiple phages to infect various pathogenes we first decided to remove D1 and D2. As we wanted to insert those two domains in the p3 of the M13 genome. Thus we use M13KO7 from New England BioLab. '''M13KO7''' is an M13 derivative which carries the mutation Met40Ile in gII , with the origin of replication from P15A and the kanamycin resistance gene from Tn903 both inserted within the M13 origin of replication.
| + | |
| − | | + | |
| − | In '''M13KO7''' we wanted to insert two restriction site (AvrII and BspI) which are compatible with XbaI and AgeI. Thus, we create two types of biobrick, one with the signal sequence of M13, and the other one with D1 and D2 of another p3 from another filamentous phages.
| + | |
| − | | + | |
| − | In our design we wanted to keep the signal sequence and D3 of M13, because their are crucial for the formation of the phage. We just want to insert D1 and D2 from another phages (we’ll call it X).
| + | |
| − | | + | |
| − | [[File:T--Aix-Marseille--P3map.png|700px|center]]
| + | |
| − | | + | |
| − | Another way to engineered M13, is to remove entierely the protein III from the phage genome and to reconstruct it in another plasmid. Thus, we create another part : p3_D3, which is the domain involved in the assembly and release of M13 particles.
| + | |
| − | | + | |
| − | ===Attachment protein===
| + | |
| − | | + | |
| − | Our goal is to create a engineered M13 phage that will be specific to an other bacteria. Thus we started to look in the bibliography and in the NCBI data base, filamentous phages that were able to infect various pathogens. D3 and the signal sequence are both the best conserved part from the attachment protein. So with protein global alignment (Needleman-Wunsch alignment), from two or three sequence at one time, we were eventually able to determinate D1 and D2.
| + | |
| − | | + | |
| − | {|
| + | |
| − | ! scope="col" |Pathogene
| + | |
| − | ! scope="col" |Filamentous phage
| + | |
| − | ! scope="col" |gene ID
| + | |
| − | |-
| + | |
| − | |''Escherichia coli''
| + | |
| − | |M13 (fd,ff)<ref name=Smeal>Smeal, S. W., Schmitt, M. A., Pereira, R. R., Prasad, A. & Fisk, J. D. Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation. Virology 500, 259–274 (2017).</ref>
| + | |
| − | |927334
| + | |
| − | |-
| + | |
| − | |''Neisseria gonorrheae''
| + | |
| − | |NgoΦ6<ref>Piekarowicz, A. et al. Neisseria gonorrhoeae Filamentous Phage NgoΦ6 Is Capable of Infecting a Variety of Gram-Negative Bacteria. J Virol 88, 1002–1010 (2014).</ref>
| + | |
| − | |1260906
| + | |
| − | |-
| + | |
| − | |''Pseudomonas aeruginosa''
| + | |
| − | |Pf3<ref>Luiten, R. G., Schoenmakers, J. G. & Konings, R. N. The major coat protein gene of the filamentous Pseudomonas aeruginosa phage Pf3: absence of an N-terminal leader signal sequence. Nucleic Acids Res 11, 8073–8085 (1983).</ref>
| + | |
| − | | + | |
| − | |1260906
| + | |
| − | |-
| + | |
| − | | rowspan="2" | ''Ralstonia solanacearum''
| + | |
| − | |RSM1Φ<ref name="T,K">T, K. et al. Genomic characterization of the filamentous integrative bacteriophages {phi}RSS1 and {phi}RSM1, which infect Ralstonia solanacearum., Genomic Characterization of the Filamentous Integrative Bacteriophages φRSS1 and φRSM1, Which Infect Ralstonia solanacearum. J Bacteriol 189, 189, 5792, 5792–5802 (2007).</ref>
| + | |
| − | |5179368
| + | |
| − | |-
| + | |
| − | |RSS1Φ<ref name="T,K">T, K. et al. Genomic characterization of the filamentous integrative bacteriophages {phi}RSS1 and {phi}RSM1, which infect Ralstonia solanacearum., Genomic Characterization of the Filamentous Integrative Bacteriophages φRSS1 and φRSM1, Which Infect Ralstonia solanacearum. J Bacteriol 189, 189, 5792, 5792–5802 (2007).</ref>
| + | |
| − | |4525385
| + | |
| − | |-
| + | |
| − | | rowspan="3" | ''Vibrio Cholerea''
| + | |
| − | |CTXΦ<ref name="Heilpern">Heilpern, A. J. & Waldor, M. K. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185, 1037–1044 (2003).</ref>
| + | |
| − | |26673076
| + | |
| − | |-
| + | |
| − | |VFJΦ(fs2)<ref>Ikema, M. & Honma, Y. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144, 1901–1906 (1998).</ref>
| + | |
| − | | + | |
| − | |1261866
| + | |
| − | |-
| + | |
| − | |VGJΦ<ref>Campos, J. et al. VGJφ, a Novel Filamentous Phage of Vibrio cholerae, Integrates into the Same Chromosomal Site as CTXφ. J. Bacteriol. 185, 5685–5696 (2003).</ref>
| + | |
| − | |1260523
| + | |
| − | |-
| + | |
| − | |''Xanthomonas campestris''
| + | |
| − | |ΦLf<ref>Tseng, Y.-H., Lo, M.-C., Lin, K.-C., Pan, C.-C. & Chang, R.-Y. Characterization of filamentous bacteriophage ΦLf from Xanthomonas campestris pv. campestris. Journal of general virology 71, 1881–1884 (1990).</ref>
| + | |
| − | |3730653
| + | |
| − | |-
| + | |
| − | |''Xanthomonas fucans''
| + | |
| − | |XacF1<ref>Ahmad, A. A., Askora, A., Kawasaki, T., Fujie, M. & Yamada, T. The filamentous phage XacF1 causes loss of virulence in Xanthomonas axonopodis pv. citri, the causative agent of citrus canker disease. Front. Microbiol. 5, (2014).</ref>
| + | |
| − | | + | |
| − | |17150318
| + | |
| − | |-
| + | |
| − | |''Xylella fastidiosa''
| + | |
| − | |XfasM23<ref>Chen, J. & Civerolo, E. L. Morphological evidence for phages in Xylella fastidiosa. Virology Journal 5, 75 (2008).</ref>
| + | |
| − | |6203562
| + | |
| − | |}
| + | |
| − | | + | |
| − | Table showing the attachment proteins from various filamentous phages.
| + | |
| − | | + | |
| − | ===Signal sequence===
| + | |
| − | The signal sequence is crucial for the excretion of p3 in the periplasm.<ref name="Heilpern">Heilpern, A. J. & Waldor, M. K. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185, 1037–1044 (2003).</ref> As we remove it with our construction, we must put another one. We choose to use the one coming from M13 as we use E. coli to produce our phage. In order to be functional, the signal peptide must be cut down from the rest of the protein. Thus, we must add the cleavage site. Using the logiciel SignalP 4.1, we saw that the cleavage is made between the alanine and the glutamate.
| + | |
| − | | + | |
| − | [[File:T--Aix-Marseille--M13pIII-Sequencesignal.jpeg|400px|center]] | + | |
| − | | + | |
| − | In order to gain flexibility, which will help the enzyme to cleave the signal sequence, we add two glycine and one serine residue which we retrotranslate, with the codon biais of E. coli K12.
| + | |
| − | | + | |
| − | The signal sequence and D1-D2 sequence are designed to make fusion protein, thus we choose to make them Freiburg assembly standard with Rfc25 prefix and sufix. This will be helpful in order to assemble our biobrick.
| + | |
| − | | + | |
| − | ===Phagemid=== | + | |
| − | | + | |
| − | As M13KO7 genome is not capable to be used for the phage assemblage, we will use a phagemid which will carry a toxin to assemble our ingeneered M13 phage.
| + | |
| − | | + | |
| − | We thought about using the Super Nova toxin (BBa_K1491017), made by the iGEM team Carnegie Mellon in 2014. This toxin has multiple benefits, because of the production of ROS occuring only in a yellow - orange light, we can produce phages-like particules carring this toxin without killing ''E.coli''. However, when this toxin will be produced in ''X. fastidiosa'' with the light coming from the sun, the bacterium will be harmed, even if it is in the Xylem vessels.
| + | |
| − | To optimise the production of this toxin in ''X. fastidiosa'' we tried to find a strong and constitutive promoter of this bacterium.
| + | |
| − | | + | |
| − | We thought about multiple ways to engineer our phagemid.
| + | |
| − | | + | |
| − | [[File:T--Aix-Marseille--pbluescript-SN.jpeg|400px|left]]
| + | |
| − | | + | |
| − | [[File:T--Aix-Marseille--pSB1C3-SN.jpeg|400px|centre]]
| + | |
| − | | + | |
| − | To deliver our toxin, either we created a phagemid that contain the oriM13 (BBa_K1445000) which will gives it the opportunity to be used in the phage construction, or we used the phagemid pBluescript II KS(+).
| + | |
| − | | + | |
| − | Both of those phagemid contain a M13 origin of replication and a gene for antibiotic resistance. We insert in both, a ''E.coli'' or ''X. fastidiosa'' promoter along with SuperNova gene.
| + | |
| − | | + | |
| − | | + | |
| − | ===''X. fastidiosa'' promoter=== | + | |
| − | | + | |
| − | | + | |
| − | [[File:T--Aix-Marseille--M13pIII-SoftBerry.jpeg|400px|right]]
| + | |
| − | | + | |
| − | Firstly, we found best bidirectional hit (BBH) between Escherichia coli str. K-12 substr. MG1655 genes and ''Xylella fastidiosa'' 9a5c ones. In order to have a strong constitutive promoter we look at highly expressed genes from ''E.coli''.<ref>S, K., J, M., A, C. & D, K. Characterizations of highly expressed genes of four fast-growing bacteria., Characterizations of Highly Expressed Genes of Four Fast-Growing Bacteria. J Bacteriol 183, 183, 5025, 5025–5040 (2001).</ref>
| + | |
| − |
| + | |
| − | Secontly, with the tool rsat, for each gene selected we take the upstream sequence from the previous gene to the ATG And with the tool BPROM we choose the sequence with predicted box with the best score. We choose XF_RS01885 which is the BBH of purA, which code for an adenylosuccinate synthetase.
| + | |
| − | | + | |
| − | Finaly, we tried to find the ribosome binding site (RBS) consensus in ''Xylella fastidiosa''. To do so we search for the anti-Shine dalgarno sequence with ''Xylella fastidiosa'' 16S ribosomal RNA gene (accession number : NR_041779). The consus found is : AGGAGG. The RBS is supposed to be 6 to 12 nucleotide upstream the ATG. So we modified the sequence. And we added Rfc10 prefix and suffix region.
| + | |
| | | | |
| | ==References== | | ==References== |
| | | | |
| | <references/> | | <references/> |
Engineering bacteriophages
Phage-like particles process.
Bacteriophages play a special role in the development of nanoscale cargo-delivery because they can be regarded as naturally occurring nanomaterials.
We chose to explore Phage-like particles (PLPs)[1] for use against pathogenic bacteria like Xylella fastidiosa.
Our goal is to use a phage-like M13 to inject a lethal toxin into the bacterium.
To help our research we asked Mireille Ansaldi, a local phage expert.
Phage systems, like M13, have been widely employed in biotechnology,
most prominently in the identification and selection of high affinity molecules through phage display [2].
The use of phages in materials science and nanotechnology is mainly due to their nanoscale size and simple life cycles.
We decided to profit from this wide application in order to target X. fastidiosa [3].
Phage-like particle design
Bacteriophages are able to generate new copies of themselves using the information in their genome [4].
In our design, we decided to limit the phage's ability to reproduce in order to contain it, as our interview with Jacques VAN HELDEN indicated this was an important point.
There are already well-tried methods to reduce the reproductive capacity of phages as well as to make phages with foreign proteins[1].
Phage-like particles (PLPs) mimic the organization of native phages but lack the viral genome.
They have been used as vehicles for drug and gene delivery and also as tools in nanobiotechnology [5].
In this project, we create M13 phage-like particles that inject toxin genes into target bacteria.
Phage-like particle in the environment
Beyond efficiency, we want our product to be environmentally friendly so that it doesn't contaminate nature with GMO's.
To investigate environmental impact, we used soil samples to test the longevity of our phages in nature.
We are also working with the [http://biam.cea.fr/drf/biam/Pages/laboratoires/lgbp.aspx LGBP lab], where we test our phage in the model organism Arabidopsis thaliana.
-
 Phage design
Phage design
-
 Environmental test
Environmental test
References
- ↑ 1.0 1.1 Roldão, A., Silva, A. C., Mellado, M. C. M., Alves, P. M. & Carrondo, M. J. T. Viruses and Virus-Like Particles in Biotechnology: Fundamentals and Applications. in Reference Module in Life Sciences (Elsevier, 2017).
- ↑ Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).
- ↑ Buttimer, C. et al. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 8, (2017).
- ↑ Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).
- ↑ Czapar, A. E. & Steinmetz, N. F. Plant viruses and bacteriophages for drug delivery in medicine and biotechnology. Current Opinion in Chemical Biology 38, 108–116 (2017).




