|
|
| (19 intermediate revisions by 5 users not shown) |
| Line 1: |
Line 1: |
| − | {{Aix-Marseille|title=Quorum sensing|toc=__TOC__}} | + | {{Aix-Marseille|title=Quorum sensing|toc=__noTOC__}} |
| | | | |
| − | Quorum sensing is a mechanism that allows bacteria to coordinate the regulation of their genes according to the density of the bacterial population<ref>Rutherford, S. T. & Bassler, B. L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb Perspect Med 2, a012427 (2012).</ref>. It allows bacteria to adopt collective patterns of gene regulation to have, at a population level, an advantageous phenotype. Quorum sensing allows [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']] to produce a biofilm, a sticky extracellular matrix composed of DNA, proteins and polysaccharides, which is one of the key problems in the plant disease because it blocks xylem vessels. | + | Quorum sensing is a mechanism that allows bacteria to coordinate their behaviour depending on the bacterial population density <ref>Rutherford, S. T. & Bassler, B. L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb Perspect Med 2, a012427 (2012).</ref>. |
| | + | It allows bacteria to adopt collective patterns of gene regulation to have, at a population level, an advantageous phenotype. |
| | + | Quorum sensing allows [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']] to produce a biofilm, a sticky extracellular matrix composed of DNA, proteins and polysaccharides, which is one of the key problems in the plant disease because it blocks xylem vessels. |
| | | | |
| | [[File:T--Aix-Marseille--QSexp.png|500px|right|thumb| Utilization of the DSF 2-cis-decenoic acid to stop the biofilm formation of bacteria.]] | | [[File:T--Aix-Marseille--QSexp.png|500px|right|thumb| Utilization of the DSF 2-cis-decenoic acid to stop the biofilm formation of bacteria.]] |
| | | | |
| − | In order to produce this biofilm, to communicate and coordinate the gene expression of the whole colony, [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] mainly uses Diffusible Signal Factor (DSF) which are fatty acids with an unsaturation in 2cis position<ref>Ionescu, M. et al. Promiscuous Diffusible Signal Factor Production and Responsiveness of the Xylella fastidiosa Rpf System. mBio 7, e01054–16 (2016).</ref>. The same route of communication also regulates virulence. In addition to quorum sensing, bacteria can also use a similar mechanism called quorum quenching to blur the quorum sensing of other bacterial populations and outcompete them. Therefore, to stop [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] to form the biofilm that causes the death of plants we choose to quench it quorum sensing activity. | + | In order to produce this biofilm, to communicate and coordinate the gene expression of the whole colony, [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] mainly uses a Diffusible Signal Factor (DSF) which is a fatty acid with a 2-cis unsaturation<ref>Ionescu, M. et al. Promiscuous Diffusible Signal Factor Production and Responsiveness of the ''Xylella fastidiosa'' Rpf System. mBio 7, e01054–16 (2016).</ref>. |
| | + | The same system of communication regulates virulence. |
| | + | In addition to quorum sensing, bacteria can also use a similar mechanism called quorum quenching to modify the quorum sensing of other bacterial populations and outcompete them. |
| | + | Therefore, to stop [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] forming the biofilm that kills plants we choose to quench it quorum sensing activity. |
| | | | |
| − | ''Pseudomonas aeruginosa'' under stress produces a specific fatty acid named 2-cis-decenoic acid, that has the ability to stop the biofilm production of many organisms<ref>Amari, D. T., Marques, C. N. H. & Davies, D. G. The Putative Enoyl-Coenzyme A Hydratase DspI Is Required for Production of the ''Pseudomonas aeruginosa'' Biofilm Dispersion Autoinducer cis-2-Decenoic Acid. J. Bacteriol. 195, 4600–4610 (2013).</ref>. Thus, we thought about using this specific DSF to stop [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']]. | + | ''Pseudomonas aeruginosa'' under stress produces a similar fatty acid named 2-cis-decenoic acid, that is able to stop biofilm formation by many organisms<ref>Amari, D. T., Marques, C. N. H. & Davies, D. G. The Putative Enoyl-Coenzyme A Hydratase DspI Is Required for Production of the ''Pseudomonas aeruginosa'' Biofilm Dispersion Autoinducer cis-2-Decenoic Acid. J. Bacteriol. 195, 4600–4610 (2013).</ref>. |
| | + | Thus, we thought of including this specific DSF in '''KILL XYL''' to stop [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] biofilm formation. |
| | | | |
| | ==Design== | | ==Design== |
| Line 13: |
Line 19: |
| | [[File:T--Aix-Marseille--pQS2.png|450px|right|thumb]] | | [[File:T--Aix-Marseille--pQS2.png|450px|right|thumb]] |
| | | | |
| − | As we wanted to limit the number of GMOs in our product we wanted to produce the 2-cis-decenoic acid in ''E. coli'' and an efficient purification system. We choose to not use ''P. aeruginosa'' because of it pathogenicity. | + | As we wanted to limit the number of GMOs in our product we wanted to produce and purify the 2-cis-decenoic acid in ''E. coli''. |
| | + | We choose to not use ''P. aeruginosa'' because of it pathogenicity. |
| | | | |
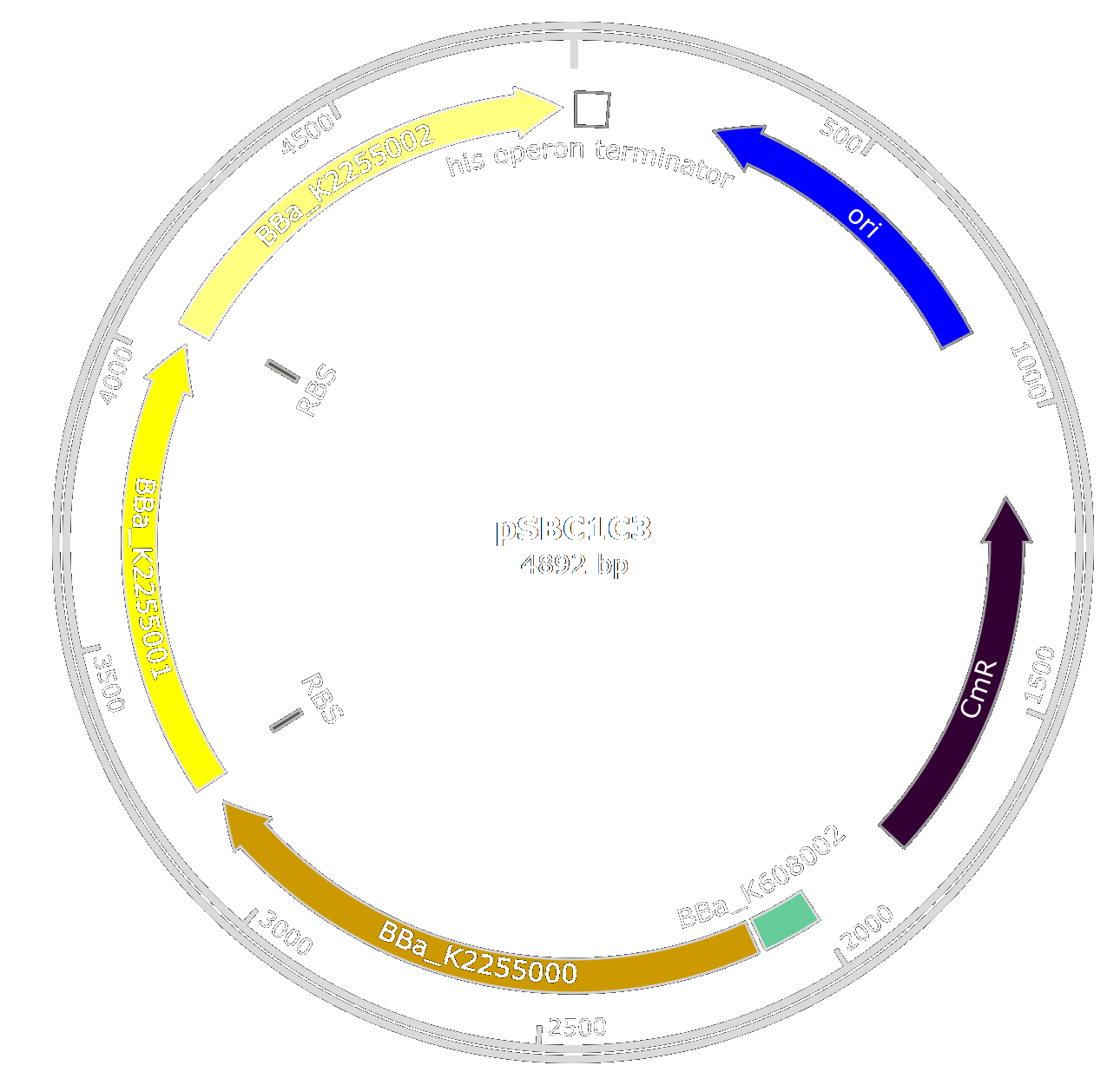
| − | Several enzymes are necessary for the production of 2-cis-decenenoic acid by ''Pseudomonas aeruginosa'', not every of them is present in ''E. coli''. Thus, to produce the fatty acid, we design the three enzymes lacking in ''E. coli'' for the production of 2-cis-decenenoic and we optimized their sequences for a production in this bacteria. Therefore we design : [http://parts.igem.org/Part:BBa_K2255000 BBa_K2255000], [http://parts.igem.org/Part:BBa_K2255001 BBa_K2255001] and [http://parts.igem.org/Part:BBa_K2255002 BBa_K2255002] | + | Several enzymes are necessary for the production of 2-cis-decenoic acid by ''Pseudomonas aeruginosa'', thay are not all present in ''E. coli''. |
| − | | + | Thus, to produce the fatty acid, we designed biobricks to produce the three enzymes lacking in ''E. coli'' for the production of 2-cis-decenenoic and we optimized their sequences for production in ''E. coli''. |
| − | As we wanted to optimize the production we design these parts to form an operon when assembled. Thus, [http://parts.igem.org/Part:BBa_K2255001 BBa_K2255001] and [http://parts.igem.org/Part:BBa_K2255002 BBa_K2255002] has a Ribosome Binding Site (RBS) integrated into the biobrick.
| + | Part [http://parts.igem.org/Part:BBa_K2255000 BBa_K2255000] is an enoyl-CoA hydratase, [http://parts.igem.org/Part:BBa_K2255001 BBa_K2255001] an acyl-CoA isomerase and [http://parts.igem.org/Part:BBa_K2255002 BBa_K2255002] a thioesterase. |
| − | | + | |
| − | Finally, to produce these enzymes in huge amounts, we decided to add a strong and constitutive promoter in E.coli ([http://parts.igem.org/Part:BBa_K608002 BBa_K608002]).
| + | |
| | | | |
| | + | We want to optimize the production of the different proteins so we designed these parts so they can be assembled to form an operon. Thus, [http://parts.igem.org/Part:BBa_K2255001 BBa_K2255001] and [http://parts.igem.org/Part:BBa_K2255002 BBa_K2255002] have a Ribosome Binding Site (RBS) integrated into the biobrick. |
| | + | To produce large amounts of these enzymes, we decided to add a strong and constitutive promoter in ''E.coli'' ([http://parts.igem.org/Part:BBa_K608002 BBa_K608002]). |
| | | | |
| | ==Results== | | ==Results== |
| | | | |
| − | As a proof of concept, we study the fatty acid effect on X. campestris. Also, we used differents experimental conditions, such as concentration of product and type of tube. After making growth bacteria in medium containing the product or not, we measured the effect of it on biofilm production with crystal violet ([https://2017.igem.org/Team:Aix-Marseille/Notebook#Biofilm_quantification Protocol]). Biofilm production can be quantified with TECAN and is proportional with absorbance. We measured fatty acid effect by adding it at the start of growth. Then we controlled the production after 24,48,72,96 hours. We also want to observe an effect after 24 hours of growth. | + | [[File:T--Aix-Marseille--biofilmtubeacide.jpeg|thumb|left|Biofilm production without fatty acid after 24, 48, 72, and 96 hours.]] |
| | + | [[File:T--Aix-Marseille--biofilmtubeacideeffet.jpeg|thumb|left|Biofilm production with fatty acid 24, 48, 72, and 96 hours.]] |
| | + | As a proof of concept, we studied the effect of the fatty acid on ''X. campestris''. |
| | + | We used several different experimental conditions, such as the concentration of product and type of tube. |
| | + | After growing bacteria in media containing the product, we measured the effect of it on biofilm production using crystal violet visually ([https://2017.igem.org/Team:Aix-Marseille/Notebook#Biofilm_quantification Protocol]). |
| | + | Biofilm production can also be quantified with TECAN and is proportional with absorbance. |
| | + | We measured fatty acid effect by adding it at the start of growth. |
| | + | Then we observed biofilm production after 24, 48, 72, and 96 hours. |
| | | | |
| − | [[File:T--Aix-Marseille--biofilmtubeacide.jpeg|thumb|left|Biofilm production without fatty acid]]
| + | These results are very encouraging. We can see a significant decrease of biofilm production after 48, 72 and 96 hours. |
| − | [[File:T--Aix-Marseille--biofilmtubeacideeffet.jpeg|thumb|left|Biofilm production with fatty acid]]
| + | However, they need to be reproduced and the biofilm quantified and studied in more detail. |
| | | | |
| | + | <!-- This does not help understanding numbers with no explanation and according to the figure 24 hours is not long enough |
| | | | |
| − | Results after fatty acid application seems encouraging. We can observe a small decrease of biofilm production after 48 hours and 72 hour, before the restoring of it. We decided to replicate the mesure and quantify the experiments numerically.
| + | We decided to replicate the mesure and quantify the experiments numerically. |
| | | | |
| | + | Raw Data: *** YOU SHOULD NOT PRESENT RAW DATA *** |
| | | | |
| | + | All the measures were made by Tecan Infinite® 200 at a wavelenght of 570nm. |
| | | | |
| | | | |
| | | | |
| − | | + | {| style="float:left" |
| − | | + | |- style="background-color:#808080;color:#fff;" |
| − | | + | || Hours |
| − | {| style="border-spacing:0;width:6.35cm;" | + | | align=right| 6 |
| − | |- style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | + | | align=right| 12 |
| − | || <span style="color:#ffffff;">Hours</span> | + | | align=right| 24 |
| − | | align=right| <span style="color:#ffffff;">6</span> | + | |
| − | | align=right| <span style="color:#ffffff;">24</span> | + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat1</span> | + | | Replicat1 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,97390002 | + | | 0,33360001 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 2,08780003 | + | | 0,44459999 |
| | + | | 0,78009999 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat2</span> | + | | Replicat2 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,97680002 | + | | 0,3337 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,68789995 | + | | 0,48379999 |
| | + | | 0,91189998 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat3</span> | + | | Replicat3 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,99870002 | + | | 0,57870001 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,73870003 | + | | 0,49349999 |
| | + | | 0,82230002 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Average</span> | + | | Average |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,98313336 | + | | 0,41533334 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,83813334 | + | | 0,47396666 |
| | + | | 0,8381 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Ecart-type</span> | + | | Ecart-type |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,01355888 | + | | 0,14147969 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,21770451
| + | | 0,02589061 |
| | + | | 0,06730557 |
| | |- | | |- |
| | |} | | |} |
| | | | |
| − | | + | {| style="float:left" |
| − | | + | |- style="background-color:#808080;color:#fff;" |
| − | {| style="border-spacing:0;width:8.58cm;" | + | || Hours |
| − | |- style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | + | | align=right| 6 |
| − | || <span style="color:#ffffff;">Hours</span> | + | | align=right| 12 |
| − | | align=right| <span style="color:#ffffff;">6</span> | + | | align=right| 24 |
| − | | align=right| <span style="color:#ffffff;">12</span> | + | |
| − | | align=right| <span style="color:#ffffff;">24</span> | + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat1</span> | + | | Replicat1 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,33360001
| + | | 0,32260001 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,44459999
| + | | 0,54210001 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,78009999 | + | | 1,16499996 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat2</span> | + | | Replicat2 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,3337
| + | | 0,456 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,48379999
| + | | 0,50389999 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,91189998
| + | | 0,82029998 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat3</span> | + | | Replicat3 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,57870001
| + | | 0,50999999 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,49349999
| + | | 0,40290001 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,82230002
| + | | 0,80269998 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Average</span> | + | | Average |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,41533334
| + | | 0,42953333 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,47396666
| + | | 0,48296667 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,8381
| + | | 0,92933331 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Ecart-type</span> | + | | Ecart-type |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,14147969
| + | | 0,0964627 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,02589061
| + | | 0,07192227 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,06730557
| + | | 0,20428294 |
| | |- | | |- |
| | |} | | |} |
| Line 109: |
Line 127: |
| | | | |
| | | | |
| − | {| style="border-spacing:0;width:6.435cm;" | + | |
| − | |- style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | + | |
| − | || <span style="color:#ffffff;">Hours</span> | + | |
| − | | align=right| <span style="color:#ffffff;">6</span> | + | |
| − | | align=right| <span style="color:#ffffff;">24</span> | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | The second experiment consist in adding the product not directly in the medium but after a certain time of growth. We choose to add the fatty acid in a culture of 24 hours. We observed the effect at time 6 hours and 24 hours later. |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | {| style="float:left" |
| | + | |- style="background-color:#808080;color:#fff;;" |
| | + | || Hours |
| | + | | align=right| 6 |
| | + | | align=right| 24 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat1</span> | + | | Replicat1 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,96560001
| + | | 0,97390002 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,44819999 | + | | 2,08780003 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat2</span> | + | | Replicat2 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,00259995 | + | | 0,97680002 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,79219997
| + | | 1,68789995 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat3</span> | + | | Replicat3 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,9734
| + | | 0,99870002 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,30620003
| + | | 1,73870003 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Average</span> | + | | Average |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,98053332
| + | | 0,98313336 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,51553333
| + | | 1,83813334 |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Ecart-type</span> | + | | Ecart-type |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,01950416
| + | | 0,01355888 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,24989862
| + | | 0,21770451 |
| | |- | | |- |
| | |} | | |} |
| | | | |
| − | | + | {| style="float:left" |
| − | | + | |- style="background-color:#808080;color:#fff;" |
| − | {| style="border-spacing:0;width:8.58cm;" | + | || Hours |
| − | |- style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | + | | align=right| 6 |
| − | || <span style="color:#ffffff;">Hours</span> | + | | align=right| 24 |
| − | | align=right| <span style="color:#ffffff;">6</span> | + | |
| − | | align=right| <span style="color:#ffffff;">12</span> | + | |
| − | | align=right| <span style="color:#ffffff;">24</span>
| + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat1</span> | + | | Replicat1 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,32260001
| + | | 0,96560001 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,54210001
| + | | 1,44819999 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 1,16499996
| + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat2</span> | + | | Replicat2 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,456 | + | | 1,00259995 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,50389999 | + | | 1,79219997 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,82029998
| + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Replicat3</span> | + | | Replicat3 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,50999999
| + | | 0,9734 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,40290001 | + | | 1,30620003 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,80269998
| + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Average</span> | + | | Average |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,42953333
| + | | 0,98053332 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,48296667 | + | | 1,51553333 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,92933331
| + | |
| | |- | | |- |
| − | | style="background-color:#808080;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | <span style="color:#ffffff;">Ecart-type</span> | + | | Ecart-type |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,0964627
| + | | 0,01950416 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,07192227
| + | | 0,24989862 |
| − | | align=right style="background-color:#ffffff;border:none;padding-top:0cm;padding-bottom:0cm;padding-left:0.123cm;padding-right:0.123cm;" | 0,20428294
| + | |
| | |- | | |- |
| | |} | | |} |
| | + | |
| | + | <div style="clear:both"></div> |
| | + | |
| | | | |
| | | | |
| Line 179: |
Line 208: |
| | Finally, visuals results bring to light some impact which deserve more research with different parameters. | | Finally, visuals results bring to light some impact which deserve more research with different parameters. |
| | | | |
| − | | + | --> |
| | | | |
| | ==References== | | ==References== |
Quorum sensing
Quorum sensing is a mechanism that allows bacteria to coordinate their behaviour depending on the bacterial population density [1].
It allows bacteria to adopt collective patterns of gene regulation to have, at a population level, an advantageous phenotype.
Quorum sensing allows Xylella fastidiosa to produce a biofilm, a sticky extracellular matrix composed of DNA, proteins and polysaccharides, which is one of the key problems in the plant disease because it blocks xylem vessels.

Utilization of the DSF 2-cis-decenoic acid to stop the biofilm formation of bacteria.
In order to produce this biofilm, to communicate and coordinate the gene expression of the whole colony, X. fastidiosa mainly uses a Diffusible Signal Factor (DSF) which is a fatty acid with a 2-cis unsaturation[2].
The same system of communication regulates virulence.
In addition to quorum sensing, bacteria can also use a similar mechanism called quorum quenching to modify the quorum sensing of other bacterial populations and outcompete them.
Therefore, to stop X. fastidiosa forming the biofilm that kills plants we choose to quench it quorum sensing activity.
Pseudomonas aeruginosa under stress produces a similar fatty acid named 2-cis-decenoic acid, that is able to stop biofilm formation by many organisms[3].
Thus, we thought of including this specific DSF in KILL XYL to stop X. fastidiosa biofilm formation.
Design
As we wanted to limit the number of GMOs in our product we wanted to produce and purify the 2-cis-decenoic acid in E. coli.
We choose to not use P. aeruginosa because of it pathogenicity.
Several enzymes are necessary for the production of 2-cis-decenoic acid by Pseudomonas aeruginosa, thay are not all present in E. coli.
Thus, to produce the fatty acid, we designed biobricks to produce the three enzymes lacking in E. coli for the production of 2-cis-decenenoic and we optimized their sequences for production in E. coli.
Part [http://parts.igem.org/Part:BBa_K2255000 BBa_K2255000] is an enoyl-CoA hydratase, [http://parts.igem.org/Part:BBa_K2255001 BBa_K2255001] an acyl-CoA isomerase and [http://parts.igem.org/Part:BBa_K2255002 BBa_K2255002] a thioesterase.
We want to optimize the production of the different proteins so we designed these parts so they can be assembled to form an operon. Thus, [http://parts.igem.org/Part:BBa_K2255001 BBa_K2255001] and [http://parts.igem.org/Part:BBa_K2255002 BBa_K2255002] have a Ribosome Binding Site (RBS) integrated into the biobrick.
To produce large amounts of these enzymes, we decided to add a strong and constitutive promoter in E.coli ([http://parts.igem.org/Part:BBa_K608002 BBa_K608002]).
Results

Biofilm production without fatty acid after 24, 48, 72, and 96 hours.

Biofilm production with fatty acid 24, 48, 72, and 96 hours.
As a proof of concept, we studied the effect of the fatty acid on X. campestris.
We used several different experimental conditions, such as the concentration of product and type of tube.
After growing bacteria in media containing the product, we measured the effect of it on biofilm production using crystal violet visually (Protocol).
Biofilm production can also be quantified with TECAN and is proportional with absorbance.
We measured fatty acid effect by adding it at the start of growth.
Then we observed biofilm production after 24, 48, 72, and 96 hours.
These results are very encouraging. We can see a significant decrease of biofilm production after 48, 72 and 96 hours.
However, they need to be reproduced and the biofilm quantified and studied in more detail.
References
- ↑ Rutherford, S. T. & Bassler, B. L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb Perspect Med 2, a012427 (2012).
- ↑ Ionescu, M. et al. Promiscuous Diffusible Signal Factor Production and Responsiveness of the Xylella fastidiosa Rpf System. mBio 7, e01054–16 (2016).
- ↑ Amari, D. T., Marques, C. N. H. & Davies, D. G. The Putative Enoyl-Coenzyme A Hydratase DspI Is Required for Production of the Pseudomonas aeruginosa Biofilm Dispersion Autoinducer cis-2-Decenoic Acid. J. Bacteriol. 195, 4600–4610 (2013).