| Line 8: | Line 8: | ||
<!-- Pour les mettre sur la pages : [[File:T--Aix-Marseille--NOM.png|TAILLE(XXXpx)|right/Left/center|thumb|Legende]]--> | <!-- Pour les mettre sur la pages : [[File:T--Aix-Marseille--NOM.png|TAILLE(XXXpx)|right/Left/center|thumb|Legende]]--> | ||
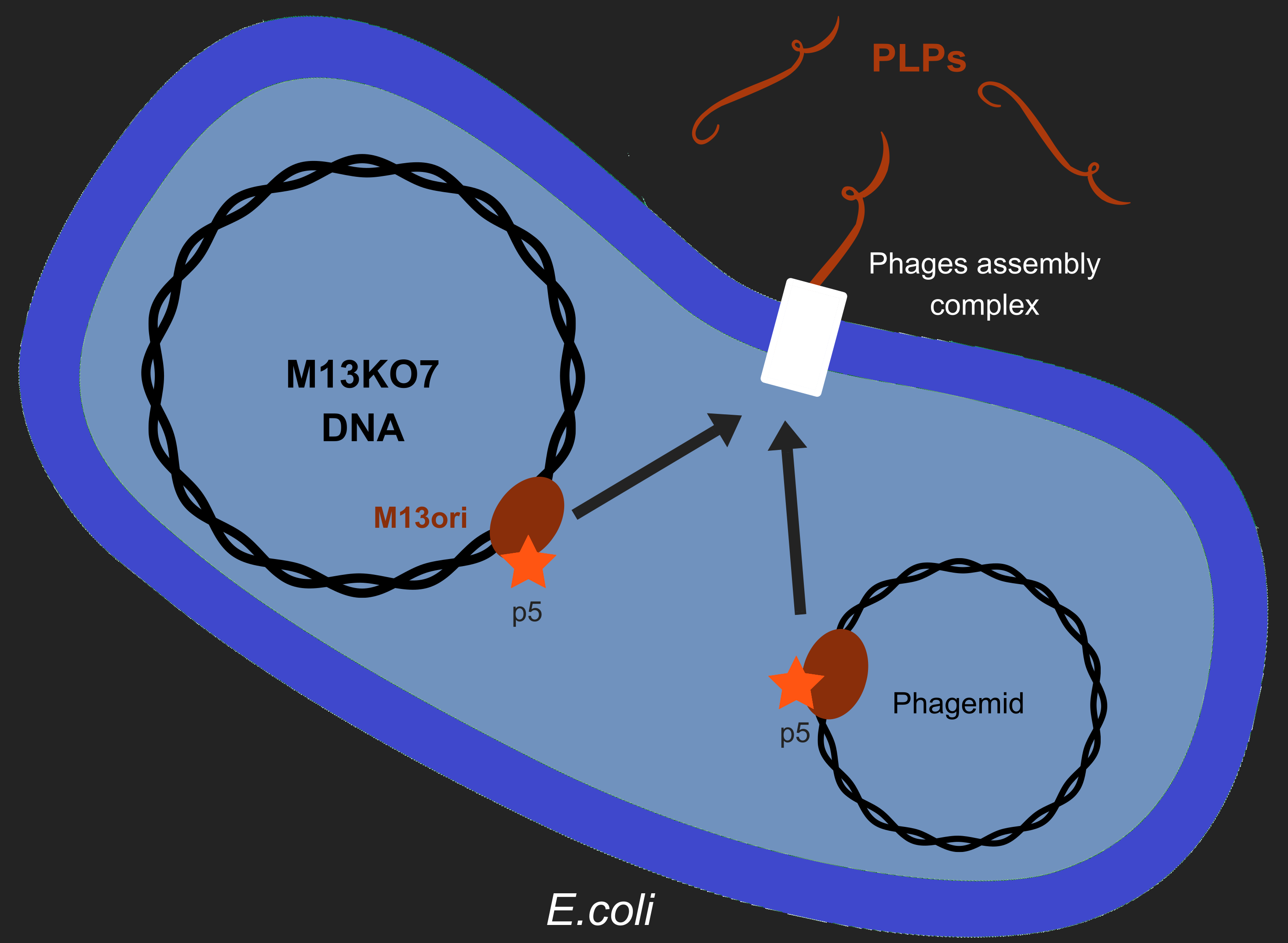
| − | [[File:T--Aix-Marseille--ModelM13.png|400px|right|thumb|In order to produce PLPs, M13KO7 (produces all the phage proteins) and the phagemid (containing the toxin gene) needs to be integrated | + | [[File:T--Aix-Marseille--ModelM13.png|400px|right|thumb|In order to produce PLPs, M13KO7 (produces all the phage proteins) and the phagemid (containing the toxin gene) needs to be integrated into the cell. During the process, if p5 takes M13KO7 it will construct a replicative phage and if it takes the phagemid it will produce PLPs.]] |
| − | Our project, [[Team:Aix-Marseille/Project|'''KILL XYL''']] will be a cure against the disease caused by the plant pathogen [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']]. One of | + | Our project, [[Team:Aix-Marseille/Project|'''KILL XYL''']] will be a cure against the disease caused by the plant pathogen [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']]. One of our approaches is to engineered [[Team:Aix-Marseille/Bacteriophages|phage-like particles]] (PLPs), that will inject toxic genes into the bacterium. Phage systems, like [[Team:Aix-Marseille/M13|M13]], have been employed in biotechnological applications, most prominently in the identification and maturation of medically-relevant binding molecules through phage display <ref>Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).</ref>. |
| − | However, production of phages as [[Team:Aix-Marseille/M13|M13]] have longterm consequences in ''E. coli'' metabolism<ref name=Smeal>Smeal ''et al'', Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017</ref>. [[Team:Aix-Marseille/M13|M13]] has a unique lifecycle among phages. One of the crucial | + | However, production of phages as [[Team:Aix-Marseille/M13|M13]] have longterm consequences in ''E. coli'' metabolism<ref name=Smeal>Smeal ''et al'', Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017</ref>. [[Team:Aix-Marseille/M13|M13]] has a unique lifecycle among phages. One of the crucial events of its multiplication is the packaging of its own DNA in the capsid. To do so, [[Team:Aix-Marseille/M13|M13]] uses it's protein 5 (p5) to target M13 origin of replication (M13ori). This step is crucial for the phage formation. |
| − | Moreover, we wanted that our phage became non-replicative to limit the spread of GMOs in the environment. That led to the creation of [[Team:Aix-Marseille/Bacteriophages|phage-like particles (PLPs)]]. The [[Team:Aix-Marseille/M13_Design|design]] of these PLPs need the genome of M13 modified to stop the replication and the addition of a phagemid in the factory cell ''E.coli''. Therefore, to create PLPs we use a modified M13 genome (M13KO7) that has several | + | Moreover, we wanted that our phage became non-replicative to limit the spread of GMOs in the environment. That led to the creation of [[Team:Aix-Marseille/Bacteriophages|phage-like particles (PLPs)]]. The [[Team:Aix-Marseille/M13_Design|design]] of these PLPs need the genome of M13 modified to stop the replication and the addition of a phagemid in the factory cell ''E.coli''. Therefore, to create PLPs we use a modified M13 genome (M13KO7) that has several genes inserts in it M13ori and we integrate a phagemid with a M13ori. |
Thanks to [[Team:Aix-Marseille/HP/Interviews|Jacques VAN HELDEN interview]] we put attention to the risk of recombination between M13KO7 and the phagemid. This question matter because we aim to create PLPs that aren't able to replicate and thus spread in the environment. Therefore, we test this possibility along our [[Team:Aix-Marseille/M13_test|wetlab tests]]. | Thanks to [[Team:Aix-Marseille/HP/Interviews|Jacques VAN HELDEN interview]] we put attention to the risk of recombination between M13KO7 and the phagemid. This question matter because we aim to create PLPs that aren't able to replicate and thus spread in the environment. Therefore, we test this possibility along our [[Team:Aix-Marseille/M13_test|wetlab tests]]. | ||
| − | Here we wanted to answer a single question : '''As p5 | + | Here we wanted to answer a single question: '''As p5 recognized M13ori, will the bacterium produces only PLPs or will it also produces replicative phages ?'''. |
==First iteration of the model== | ==First iteration of the model== | ||
| − | We started with an excessively simple model to see | + | We started with an excessively simple model to see whether our understanding of our system was profound enough to allow room for such simplifications. We made the following assumptions in this spirit: |
Which led to the following model: | Which led to the following model: | ||
| Line 52: | Line 52: | ||
==Second iteration== | ==Second iteration== | ||
| − | <!--les réference se place entre <ref>xxx</ref>. Si tu souhaite mettre plusieur | + | <!--les réference se place entre <ref>xxx</ref>. Si tu souhaite mettre plusieur fois la meme ref il faut que tu mettes <ref name=nom>xxx</ref>. Sinon il va croire que c'est des ref differentes--> |
| − | This time, we decided to | + | This time, we decided to adopt the model of a "wild-type" M13's biology <ref name=Smeal>Smeal ''et al'', Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017</ref><ref>Smeal ''et al'', Simulation of the M13 life cycle II: Investigation of the control mechanisms of M13 infection and establishment of the carrier state, Virology, 500, January 2017</ref> to fit our own engineered version of M13 and better inform wet lab experiments, as well as explain some of their preliminary findings! |
=Model validation= | =Model validation= | ||
| − | compare particle profile to that obtained in | + | compare particle profile to that obtained in wet lab |
= --> Insufficient p3= | = --> Insufficient p3= | ||
Revision as of 15:26, 29 October 2017
Engineered M13 modelling
Contents
Our project, KILL XYL will be a cure against the disease caused by the plant pathogen Xylella fastidiosa. One of our approaches is to engineered phage-like particles (PLPs), that will inject toxic genes into the bacterium. Phage systems, like M13, have been employed in biotechnological applications, most prominently in the identification and maturation of medically-relevant binding molecules through phage display [1].
However, production of phages as M13 have longterm consequences in E. coli metabolism[2]. M13 has a unique lifecycle among phages. One of the crucial events of its multiplication is the packaging of its own DNA in the capsid. To do so, M13 uses it's protein 5 (p5) to target M13 origin of replication (M13ori). This step is crucial for the phage formation.
Moreover, we wanted that our phage became non-replicative to limit the spread of GMOs in the environment. That led to the creation of phage-like particles (PLPs). The design of these PLPs need the genome of M13 modified to stop the replication and the addition of a phagemid in the factory cell E.coli. Therefore, to create PLPs we use a modified M13 genome (M13KO7) that has several genes inserts in it M13ori and we integrate a phagemid with a M13ori.
Thanks to Jacques VAN HELDEN interview we put attention to the risk of recombination between M13KO7 and the phagemid. This question matter because we aim to create PLPs that aren't able to replicate and thus spread in the environment. Therefore, we test this possibility along our wetlab tests.
Here we wanted to answer a single question: As p5 recognized M13ori, will the bacterium produces only PLPs or will it also produces replicative phages ?.
First iteration of the model
We started with an excessively simple model to see whether our understanding of our system was profound enough to allow room for such simplifications. We made the following assumptions in this spirit:
Which led to the following model:
| Assumption | Reasonning |
|---|---|
| Neglecting cooperative effects | |
| Constant rate of p5 transcription/traduction | |
| One M13KO7 per factory cell | |
| Phage assembly isn't a limiting factor | |
| Neglecting stochasticity |
Second iteration
This time, we decided to adopt the model of a "wild-type" M13's biology [2][3] to fit our own engineered version of M13 and better inform wet lab experiments, as well as explain some of their preliminary findings!
Model validation
compare particle profile to that obtained in wet lab
--> Insufficient p3
could cause the packing of several DNA molecules per phage.
What is the ideal number of plasmid copies
Wet-lab and dry-lab back and forth
References
- ↑ Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).
- ↑ 2.0 2.1 Smeal et al, Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017
- ↑ Smeal et al, Simulation of the M13 life cycle II: Investigation of the control mechanisms of M13 infection and establishment of the carrier state, Virology, 500, January 2017