| Line 297: | Line 297: | ||
<h5> Mating switch of yeasts</h5> | <h5> Mating switch of yeasts</h5> | ||
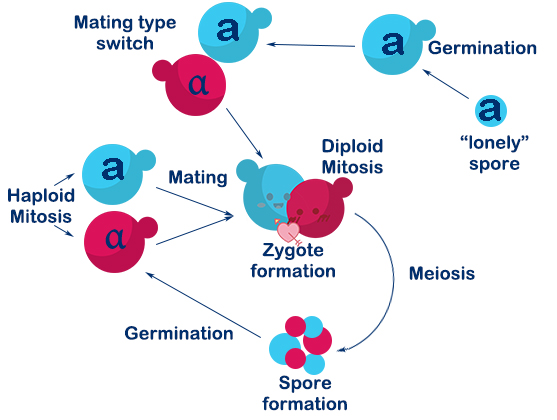
<p>Saccharomyces cerevisiae is a single-celled organism with three types, called a, α, and a/α. In Saccharomyces cerevisiae, three cell types differ from each other in their DNA content at the MAT locus which specifies the cell types. In nature, the two haploid cell types (a and α) of budding yeast are able to interconvert in a reversible manner by DNA-rearrangement with a DSB at the MAT locus, and this process is called mating-type switch.</p> | <p>Saccharomyces cerevisiae is a single-celled organism with three types, called a, α, and a/α. In Saccharomyces cerevisiae, three cell types differ from each other in their DNA content at the MAT locus which specifies the cell types. In nature, the two haploid cell types (a and α) of budding yeast are able to interconvert in a reversible manner by DNA-rearrangement with a DSB at the MAT locus, and this process is called mating-type switch.</p> | ||
| − | <p>The DSB at MAT locus is caused by HO endonuclease (a kind of site-specific endonuclease expressed by HO gene). DSBs in chromosomes can be repaired either by homologous recombination (HR) or by nonhomologous end-joining (NHEJ). In S.C haploids, the DSB caused by HO endonuclease mostly repaired by HR with HML(α) and HMR(a) as donors. If the donor is HML(α), the mating-type will become α, and vice versa. In this way, a haploid budding yeast is able to achieve mating-type switch. | + | <p>The DSB at MAT locus is caused by HO endonuclease (a kind of site-specific endonuclease expressed by HO gene). DSBs in chromosomes can be repaired either by homologous recombination (HR) or by nonhomologous end-joining (NHEJ). In S.C haploids, the DSB caused by HO endonuclease mostly repaired by HR with HML(α) and HMR(a) as donors. If the donor is HML(α), the mating-type will become α, and vice versa. In this way, a haploid budding yeast is able to achieve mating-type switch. </p> |
| + | <div class="zxx_zoom_demo" align="center"> | ||
| + | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| + | <div class="small_pic_demo" align="center"> | ||
| + | <a href="#pic_four"> | ||
| + | <img src=" https://static.igem.org/mediawiki/2017/6/6f/Tianjin-ho-design-fig4.jpeg"></a> | ||
| + | <p style="font-size:15px;text-align:center"><br/>Figure.4</p> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id="pic_four" style="display:none;"><img src=" https://static.igem.org/mediawiki/2017/6/6f/Tianjin-ho-design-fig4.jpeg"><p style="font-size:15px;text-align:center"><br/>Figure.4</p></div> | ||
| + | |||
<h5>Artificial Transcription Factors——Z4EV</h5> | <h5>Artificial Transcription Factors——Z4EV</h5> | ||
<p>Thanks to R. Scott McIsaac and Benjamin L. Oakes’s former work, we learned that Z4EV is a kind of fusion protein with three domains – DNA binding domain (DBD), estrogen receptor (ER) and VP16 activation domain. In the absence of β-estradiol, the ER interacts with Hsp90 chaperone complex and keep the ATF out of the nucleus. This AFT will provide a strong transcriptional activator that is dependent on the presence of β-estradiol. By using a synthetic 4-time-repeated zinc-finger DBD array from the mouse TF Zif268, residual off-target effects have been totally avoided. </p> | <p>Thanks to R. Scott McIsaac and Benjamin L. Oakes’s former work, we learned that Z4EV is a kind of fusion protein with three domains – DNA binding domain (DBD), estrogen receptor (ER) and VP16 activation domain. In the absence of β-estradiol, the ER interacts with Hsp90 chaperone complex and keep the ATF out of the nucleus. This AFT will provide a strong transcriptional activator that is dependent on the presence of β-estradiol. By using a synthetic 4-time-repeated zinc-finger DBD array from the mouse TF Zif268, residual off-target effects have been totally avoided. </p> | ||
Revision as of 14:17, 27 October 2017
/* OVERRIDE IGEM SETTINGS */
Design
Background
Human existence on earth is almost impossible without the heavy metals. Even though important to mankind, exposure to them during production, usage and their uncontrolled discharge in to the environment has caused lots of hazards to man, other organisms and the environment itself. Heavy metals can enter human tissues and organs via inhalation, diet, and manual handling. As the process of urbanization and industrialization goes deeper and deeper, heavy metal pollution, a noticeable threaten to almost all the creatures, has become an essential problem to solve.
According to our human practice, the situation of heavy metal pollution (copper and cadmium ions) is marked on a world map, and the severity of heavy metal pollution has been increasing all over this map. Places with serious pollution includes middle Asia, eastern Asia, southern Europe, and Latin America. In addition, not only fresh water sources, but also soil and crops are seriously contaminated by heavy metals. On average, during three out of ten suppers we have, we absorb excess heavy metals over the standard concentration.
Considering the rigorous situation we face, our team decided to design an advanced system for typical toxic heavy metal disposal based on Saccharomyces cerevisiae.