Liu Zichen (Talk | contribs) |
Liu Zichen (Talk | contribs) |
||
| Line 18: | Line 18: | ||
$('div.small_pic_big a').fancyZoom({scaleImg: true, closeOnClick: true}); | $('div.small_pic_big a').fancyZoom({scaleImg: true, closeOnClick: true}); | ||
$('div.small_pic_demo a').fancyZoom({scaleImg: true, closeOnClick: true}); | $('div.small_pic_demo a').fancyZoom({scaleImg: true, closeOnClick: true}); | ||
| + | $('div.small_pic_demo_long a').fancyZoom({scaleImg: true, closeOnClick: true}); | ||
$('div.small_pic_long a').fancyZoom({scaleImg: true, closeOnClick: true}); | $('div.small_pic_long a').fancyZoom({scaleImg: true, closeOnClick: true}); | ||
$('div.small_pic_mid a').fancyZoom({scaleImg: true, closeOnClick: true}); | $('div.small_pic_mid a').fancyZoom({scaleImg: true, closeOnClick: true}); | ||
| Line 241: | Line 242: | ||
<p>Aiming to achieve MTS for environmental use, it is essential to make sure that when the MAT locus has DSB(double strands break) cleaved by HO, our type-a (MATa) yeast can only become type-α (MATα). Therefore, we used a Ura-tag to replace the HMR(a) domain in chromosome Ⅲ. In this way the HMR will no longer be the donor for the homologous recombination in the repairing process for MAT cleavage. </p> | <p>Aiming to achieve MTS for environmental use, it is essential to make sure that when the MAT locus has DSB(double strands break) cleaved by HO, our type-a (MATa) yeast can only become type-α (MATα). Therefore, we used a Ura-tag to replace the HMR(a) domain in chromosome Ⅲ. In this way the HMR will no longer be the donor for the homologous recombination in the repairing process for MAT cleavage. </p> | ||
<p>Since the change of mating type may appear successively, there is a great possibility that the same type haploid mate with each other. To avoid the existence of meaningless mating , we built an vector to express MATα genes to produce a1-α2 stable corepressor so that the haploid will regard itself as a diploid and prevent mating unless the MATa locus changes to the other one. After selection, by homologous recombination, we deleted the Ura-tag for further usage. We selected the target colonies(SynⅩdUra) via 5Foa plates. </p> | <p>Since the change of mating type may appear successively, there is a great possibility that the same type haploid mate with each other. To avoid the existence of meaningless mating , we built an vector to express MATα genes to produce a1-α2 stable corepressor so that the haploid will regard itself as a diploid and prevent mating unless the MATa locus changes to the other one. After selection, by homologous recombination, we deleted the Ura-tag for further usage. We selected the target colonies(SynⅩdUra) via 5Foa plates. </p> | ||
| − | <div class=" | + | <div class="zxx_zoom_demo_long" align="center"> |
<script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| − | <div class=" | + | <div class="small_pic_demo_long" align="center"> |
<a href="#pic_one"> | <a href="#pic_one"> | ||
<img src=" https://static.igem.org/mediawiki/2017/f/f0/Tianjin-ho-design-fig1.jpeg"></a> | <img src=" https://static.igem.org/mediawiki/2017/f/f0/Tianjin-ho-design-fig1.jpeg"></a> | ||
| Line 265: | Line 266: | ||
<p>In this pathway, we introduced one kind of artificial transcription factor (ATF)—Z4EV into the regulation of HO gene expression. With Z4EV working with our Modified. Gal1 promoter, we hoped to reach the on off-target dynamic control of HO gene expression. | <p>In this pathway, we introduced one kind of artificial transcription factor (ATF)—Z4EV into the regulation of HO gene expression. With Z4EV working with our Modified. Gal1 promoter, we hoped to reach the on off-target dynamic control of HO gene expression. | ||
Our designing for getting the Modified. Gal1-HO-CYC1 parts (MGHC) is quite the same as that for GHC mentioned above, only that we acquired our Modified. Gal1 part from the gene synthesis. </p> | Our designing for getting the Modified. Gal1-HO-CYC1 parts (MGHC) is quite the same as that for GHC mentioned above, only that we acquired our Modified. Gal1 part from the gene synthesis. </p> | ||
| − | <div class=" | + | <div class="zxx_zoom_demo_long" align="center"> |
<script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| − | <div class=" | + | <div class="small_pic_demo_long" align="center"> |
<a href="#pic_two"> | <a href="#pic_two"> | ||
<img src=" https://static.igem.org/mediawiki/2017/0/0c/Tianjin-ho-result-real-fig2.jpeg"></a> | <img src=" https://static.igem.org/mediawiki/2017/0/0c/Tianjin-ho-result-real-fig2.jpeg"></a> | ||
| Line 281: | Line 282: | ||
<p>Next, for double using the Leu-tag, we introduce Vika/vox system. We intended to attach the Vika operator (Tdh3p-Vikc-Tdh2t, TVT) following the Z4EV gene. We had two groups of yeasts, as mentioned above, one of them aimed to accomplish MTS and becoming MATα, the other with functional genes remained as MATa. According to our design, the former will express Vika recombinase, and the other contain functional genes whose expressions are controlled by vox-Terminator-vox structure. Thus, the function gene’s expression will be initiate during the cell fusion in yeast mating process. </p> | <p>Next, for double using the Leu-tag, we introduce Vika/vox system. We intended to attach the Vika operator (Tdh3p-Vikc-Tdh2t, TVT) following the Z4EV gene. We had two groups of yeasts, as mentioned above, one of them aimed to accomplish MTS and becoming MATα, the other with functional genes remained as MATa. According to our design, the former will express Vika recombinase, and the other contain functional genes whose expressions are controlled by vox-Terminator-vox structure. Thus, the function gene’s expression will be initiate during the cell fusion in yeast mating process. </p> | ||
| − | <div class=" | + | <div class="zxx_zoom_mid" align="center"> |
<script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| − | <div class=" | + | <div class="small_pic_mid" align="center"> |
| − | <a href="# | + | <a href="#pic_hao"> |
<img src=" https://static.igem.org/mediawiki/2017/a/a6/Tianjin-ho-design-fig3.jpeg"></a> | <img src=" https://static.igem.org/mediawiki/2017/a/a6/Tianjin-ho-design-fig3.jpeg"></a> | ||
<p style="font-size:15px;text-align:center"><br/>Fig. 1-3. Z4EV and Vika. </p> | <p style="font-size:15px;text-align:center"><br/>Fig. 1-3. Z4EV and Vika. </p> | ||
| Line 291: | Line 292: | ||
</div> | </div> | ||
| − | <div id=" | + | <div id="pic_hao" style="display:none;"><img src=" https://static.igem.org/mediawiki/2017/a/a6/Tianjin-ho-design-fig3.jpeg"><p style="font-size:15px;text-align:center"><br/>Fig. 1-3. Z4EV and Vika. |
</p></div> | </p></div> | ||
<h4>TEST of MTS </h4> | <h4>TEST of MTS </h4> | ||
| Line 401: | Line 402: | ||
</p> | </p> | ||
| − | <div class=" | + | <div class="zxx_zoom_mid" align="center"> |
<script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| − | <div class=" | + | <div class="small_pic_mid" align="center"> |
<a href="#pic_fiftyseven"> | <a href="#pic_fiftyseven"> | ||
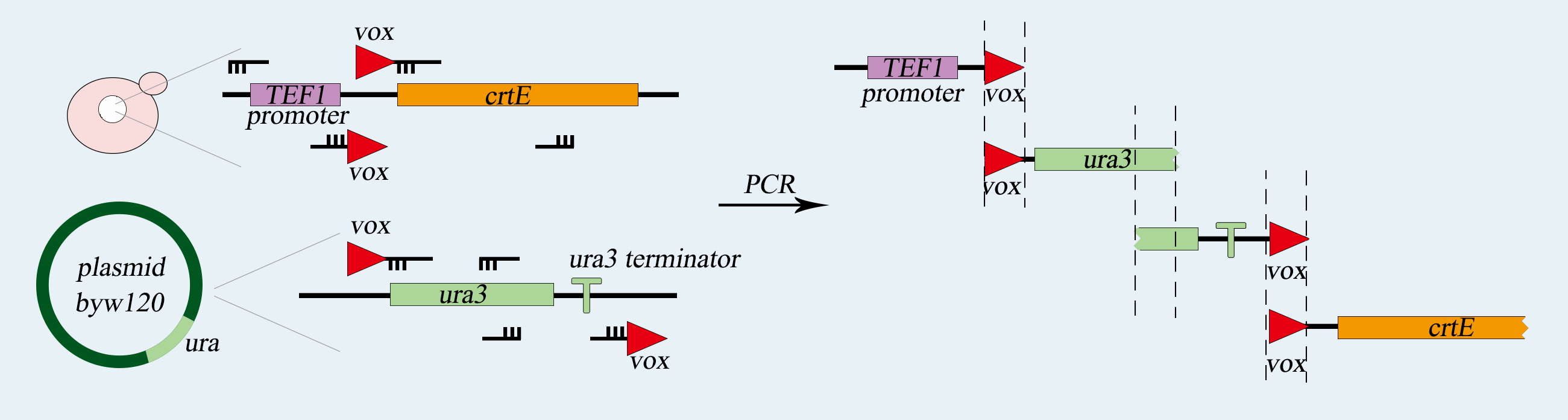
<img src="https://static.igem.org/mediawiki/2017/5/56/Tianjin-The_obtention_of_vox-ura3-terminator-vox_structure.png"></a> | <img src="https://static.igem.org/mediawiki/2017/5/56/Tianjin-The_obtention_of_vox-ura3-terminator-vox_structure.png"></a> | ||
| Line 419: | Line 420: | ||
</p> | </p> | ||
| − | <div class=" | + | <div class="zxx_zoom_mid" align="center"> |
<script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| − | <div class=" | + | <div class="small_pic_mid" align="center"> |
<a href="#pic_fiftysix"> | <a href="#pic_fiftysix"> | ||
<img src="https://static.igem.org/mediawiki/2017/9/96/Tianjin-The_obtention_of_vox-RFP-terminators-vox_structure.png"></a> | <img src="https://static.igem.org/mediawiki/2017/9/96/Tianjin-The_obtention_of_vox-RFP-terminators-vox_structure.png"></a> | ||
| Line 593: | Line 594: | ||
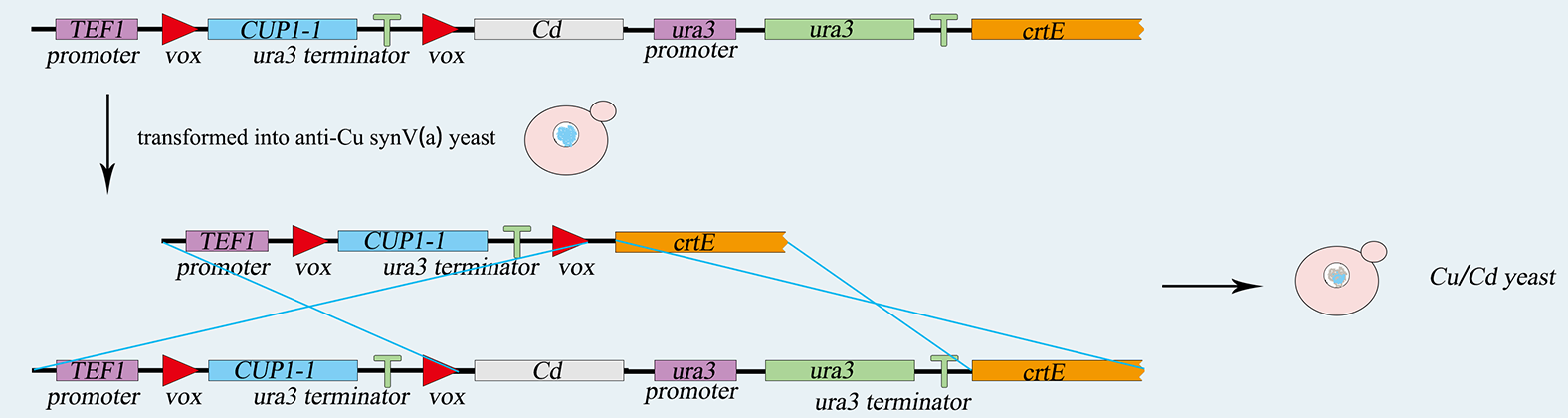
<h5>1) construction of <i>S.C-Cu(Cd)</i> </h5> | <h5>1) construction of <i>S.C-Cu(Cd)</i> </h5> | ||
<p>The <i>TEF</i> promoter, the <i>Cup1</i> gene, and the <i>Ura3</i> terminator are ligated together by overlap PCR, and then the sequence is integrated into <i>vox-ura3-vox</i> system by yeast's homologous recombination. <i>5-FOA</i> plate helps us to screen the correct cell after transferring. Similarly, the <i>LIMT</i> gene and the <i>Ura3</i> nutritional label are integrated into the same chromosome. Screened by <i>SC-ura3</i>, we complete genetic circuit as figure 5-1,5-2.showing below.</p> | <p>The <i>TEF</i> promoter, the <i>Cup1</i> gene, and the <i>Ura3</i> terminator are ligated together by overlap PCR, and then the sequence is integrated into <i>vox-ura3-vox</i> system by yeast's homologous recombination. <i>5-FOA</i> plate helps us to screen the correct cell after transferring. Similarly, the <i>LIMT</i> gene and the <i>Ura3</i> nutritional label are integrated into the same chromosome. Screened by <i>SC-ura3</i>, we complete genetic circuit as figure 5-1,5-2.showing below.</p> | ||
| − | <div class=" | + | <div class="zxx_zoom_mid" align="center"> |
<script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2017.igem.org/Team:Tianjin/Resources/JS:zoom?action=raw&ctype=text/javascript"></script> | ||
| − | <div class=" | + | <div class="small_pic_mid" align="center"> |
<a href="#pic_seventy-one"> | <a href="#pic_seventy-one"> | ||
<img src="https://static.igem.org/mediawiki/2017/2/2c/Design.cu.yeast.png"></a> | <img src="https://static.igem.org/mediawiki/2017/2/2c/Design.cu.yeast.png"></a> | ||
Revision as of 14:34, 30 October 2017
/* OVERRIDE IGEM SETTINGS */
Design
Background
Human existence on earth is almost impossible without the heavy metals. Even though important to mankind, exposure to them during production, usage and their uncontrolled discharge in to the environment has caused lots of hazards to man, other organisms and the environment itself. Heavy metals can enter human tissues and organs via inhalation, diet, and manual handling. As the process of urbanization and industrialization goes deeper and deeper, heavy metal pollution, a noticeable threaten to almost all the creatures, has become an essential problem to solve.
According to our human practice, the situation of heavy metal pollution (copper and cadmium ions) is marked on a world map, and the severity of heavy metal pollution has been increasing all over this map. Places with serious pollution includes middle Asia, eastern Asia, southern Europe, and Latin America. In addition, not only fresh water sources, but also soil and crops are seriously contaminated by heavy metals. On average, during three out of ten suppers we have, we absorb excess heavy metals over the standard concentration.
Considering the rigorous situation we face, our team decided to design an advanced system for typical toxic heavy metal disposal based on Saccharomyces cerevisiae.