| Line 26: | Line 26: | ||
<br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
| − | This figure showed that S. cerevisiae of Funazushi's growth rate at high salt concentration was faster than S. cerevisiae BY4742. | + | This figure (fig.2) showed that S. cerevisiae of Funazushi's growth rate at high salt concentration was faster than S. cerevisiae BY4742. |
===The growth of S. cerevisiae BY4742 cells and S. cerevisiae of Funazushi was tested on plates containing Lactic acid.=== | ===The growth of S. cerevisiae BY4742 cells and S. cerevisiae of Funazushi was tested on plates containing Lactic acid.=== | ||
| Line 62: | Line 62: | ||
<br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
| − | The completed PCR product of the coding region of sod2 was integrated into pYES2 plasmid and transfered | + | The completed PCR product of the coding region of sod2 was integrated into pYES2 plasmid and transfered th |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Revision as of 05:18, 1 November 2017
Nagahama
Contents
Result and Discussion
Identification of yeast of Funazushi
Funazushi was touched with a toothpick and streaked in YPD medium to grow yeast colonies. Colony PCR was performed with a primer that amplifies ITS 1 region. We sequenced the PCR products and identified yeasts of Funazushi. From the result of the sequence, the yeast of Funazushi was identified as S. cerevisiae. Therefore we decided to use S. cerevisiae BY 4742 which is commonly used in experiment as yeast to add nutrition in Funazushi.
proof of concept
First, S. cerevisiae BY4742 in order to be dominant species at high salt concentration and low pH,we investigated whether it is necessary to develop a new recombinant S. cerevisiae adapting severe environment. Accoding thesis, it is known that the salt concentration of Funazushi is 4% and pH is 3.7.[1]So we experimented whether S. cerevisiae BY 4742 could grow like S. cerevisiae of Funazushi under this severe environment.
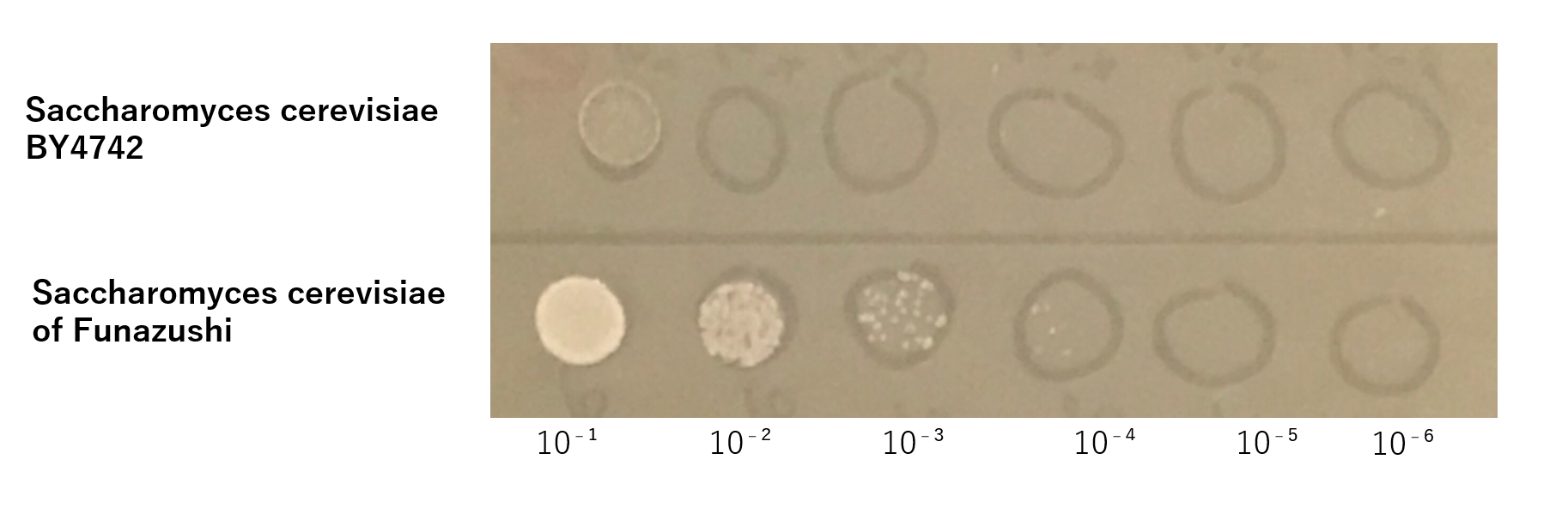
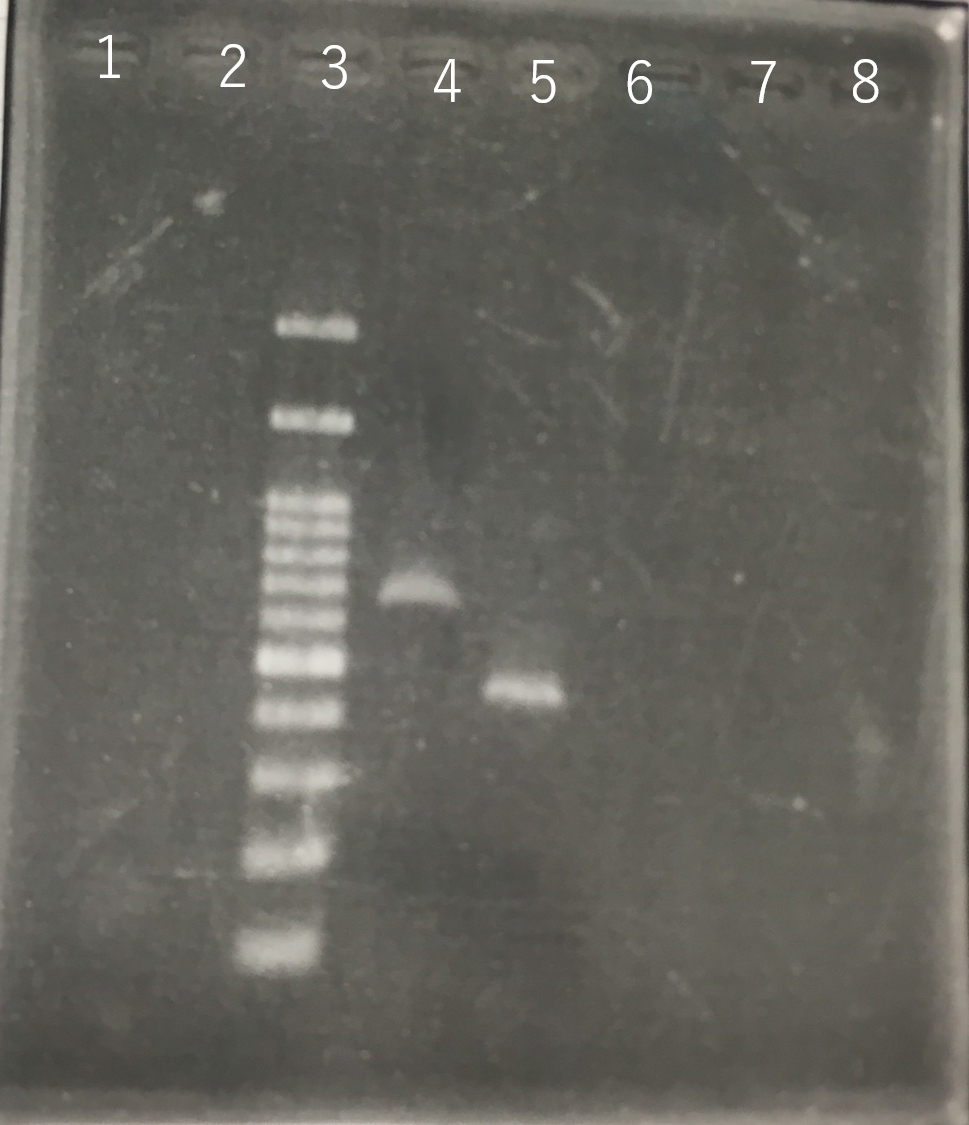
The growth of S. cerevisiae BY4742 cells and S. cerevisiae of Funazushi was tested on plates containing sodium.
This figure (fig.2) showed that S. cerevisiae of Funazushi's growth rate at high salt concentration was faster than S. cerevisiae BY4742.
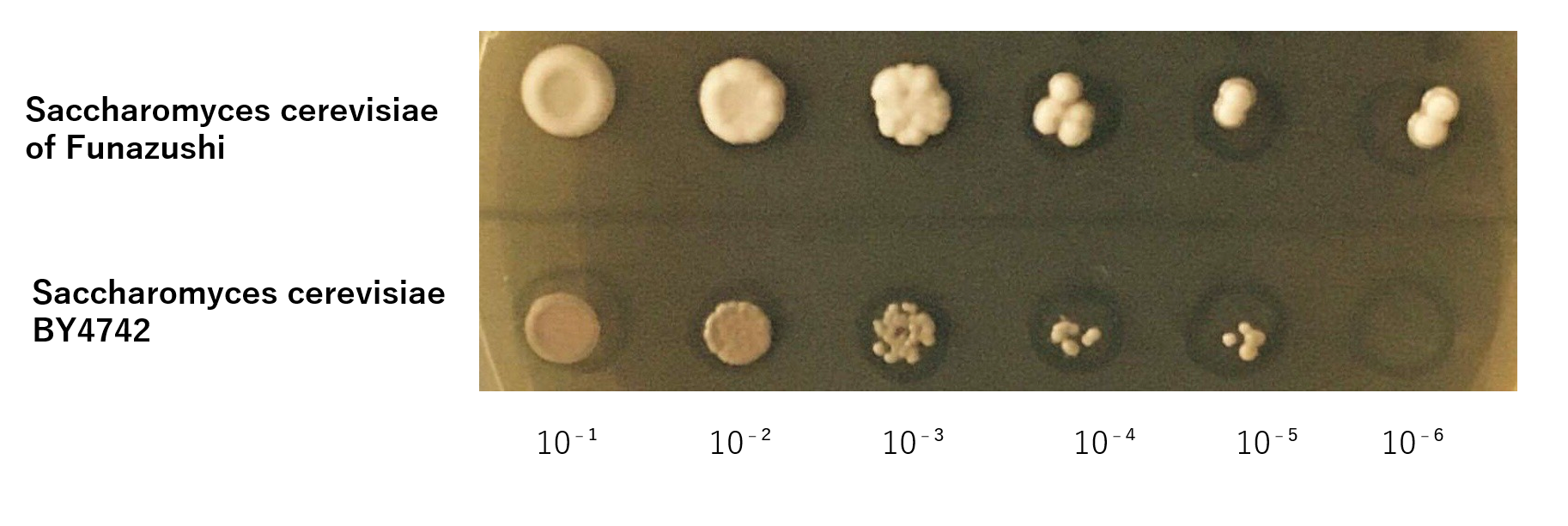
The growth of S. cerevisiae BY4742 cells and S. cerevisiae of Funazushi was tested on plates containing Lactic acid.
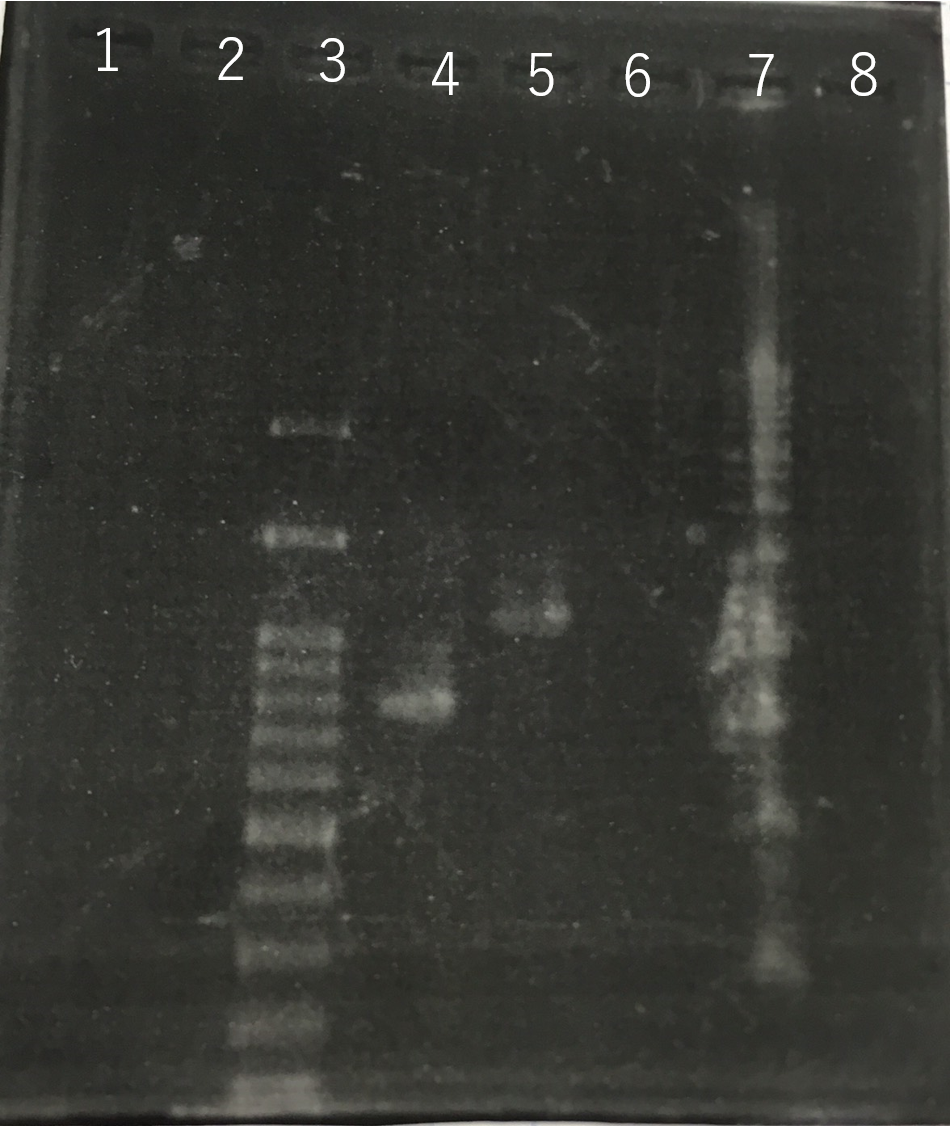
Confirmation whether sod2 were inserted into pYES2 plasmid.
The front and the back part of the intron of sod2 were separately amplified by PCR. The forward primer of sod2 located back the intron contained the sequence of reverse Primer in the front of intron as an adapter.
Using these two PCR products, fusion PCR was performed.
The completed PCR product of the coding region of sod2 was integrated into pYES2 plasmid and transfered th