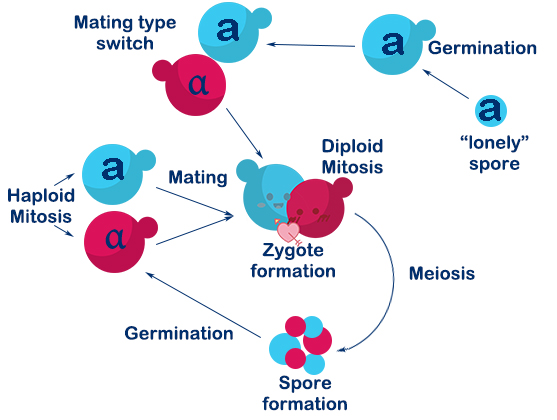
OVERVIEW
Vika-vox system is used in our project in order to switch the expression from RFP to β-carotene, as a characterization of our Mating Switcher. In this way, we can easily visualize the function of our switcher through its color, as well as measure its efficiency and error rate.
Vika-vox system mainly consists of vox sites and reporting parts. At first the expression of yEmRFP will be activating and the expression of β-carotene will be inhibited so that we can detect red fluorescence when recombinase Vika doesn’t exist in Saccharomyces cerevisiae. After the expression of recombinase Vika, with the deletion of yEmRFP and terminators flanked by vox locus, β-carotene expresses and the strains take on orange. This is the whole characterization process of Mating Switcher.
THEORETICAL BACKGROUND
1) Vika-vox System
Genome editing is emerging as a powerful technology platform which paved the way for exploring the nature of life comprehensively and systematically. Site-specific DNA recombinases have been tamed as a powerful tool in genome editing, such as Vika/vox and Cre/loxp. Site-specifc DNA recombinase Vika, originally identifed in a gram-negative bacterium Vibro coralliilyticus, could functionally and specifcally deleted genomic DNA fragment via recognizing specifc DNA site vox in yeast Saccharomyces cerevisiae and other spices, including mammal cell and bacteria.
Recently a milestone was reached with achieved total synthesis of functional chromosome Ⅴ DNA in yeast, Saccharomyces Cerevisiae. This chromosome owns lots of loxPsym sites. In spite of DNA sequence in recognizing site of loxP and vox shares a high similarity, it was demonstrated that activities of the two recombination systems were strictly independent to each other in yeast. Furthermore, the Vika/vox system functioned properly even in yeast cell carrying the synthesized chromosome on which lots of loxPsym sites are encoded.
2) Carotenogenic Pathway
Carotenoids are a class of pigments of commercial interest that have important biological functions. Certain carotenoids can be synthesized by biotechnology, by either homologous or heterologous production. One example is that β-carotene can be overexpressed in S. cerevisiae, by introduced carotenogenic genes, crtE, crtI, and crtYB, from the carotenoid-producing yeast Xanthophyllomyces dendrorhous. Like X. dendrorhous, S. cerevisiae is able to produce FPP and converts it into GGPP, the basic building block of carotenoids. Conversion of FPP into GGPP is catalyzed by GGPP synthase encoded by BTS1 in S. cerevisiae. Therefore, overexpression of only crtYB and crtI from X. dendrorhous in S. cerevisiae should generally be sufficient to transform S. cerevisiae into a β-carotene-producing organism. Additional overexpression of crtE from X. dendrorhous will increase GGPP levels and thereby enhance β-carotene production.

Fig.2-1. The carotenogenic pathway in X. dendrorhous

Fig.2-1. The carotenogenic pathway in X. dendrorhous
EXPERIMENT DESIGN
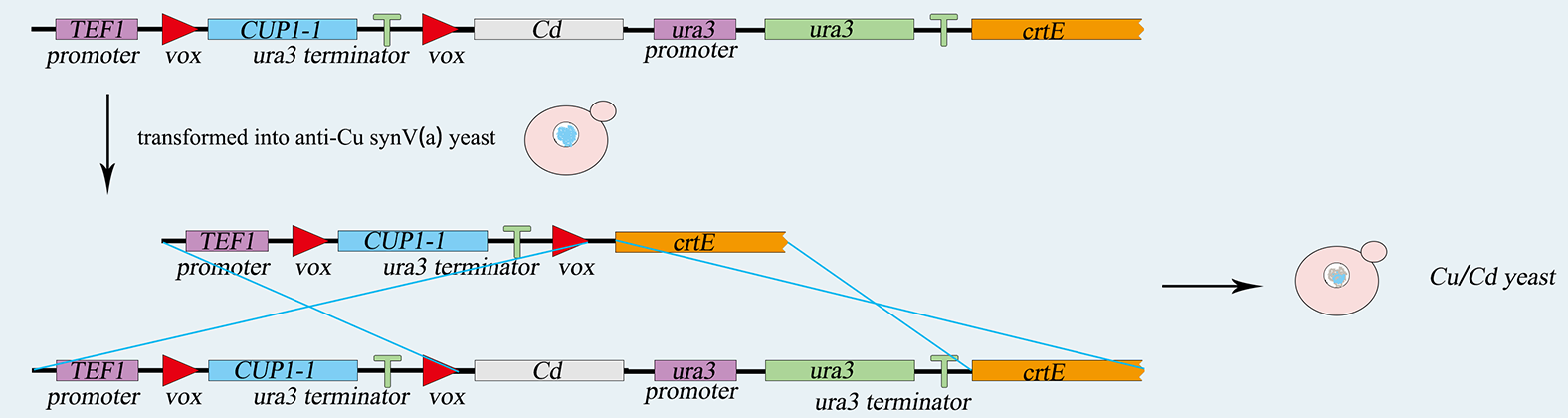
1) Construction of vox-ura3-terminator-vox Structure
We will use SynⅤ of Saccharomyces cerevisiae to load our device, which is a haploid with mating type of a. First of all, we use PCR to amplify basic parts including TEF promoter, ura3 gene, ura3-terminator and β-carotene gene. Among them, ura3 gene and ura3-terminator are flanked by vox locus. Then we use overlap PCR to combine these parts together. The next step is transform this composite part into Saccharomyces cerevisiae. We screen for the correctly transformed cell by using the Sc-Ura plate. For the purpose of verifying desired strain PVUVC, we use colony PCR to amplify the TEF promoter-vox-ura3 structure and ura3 terminator-vox-β-carotene structure. The length of the strip will be observed by agarose gel electrophoresis.

Fig.2-2. The obtention of vox-ura3-terminator-vox structure.
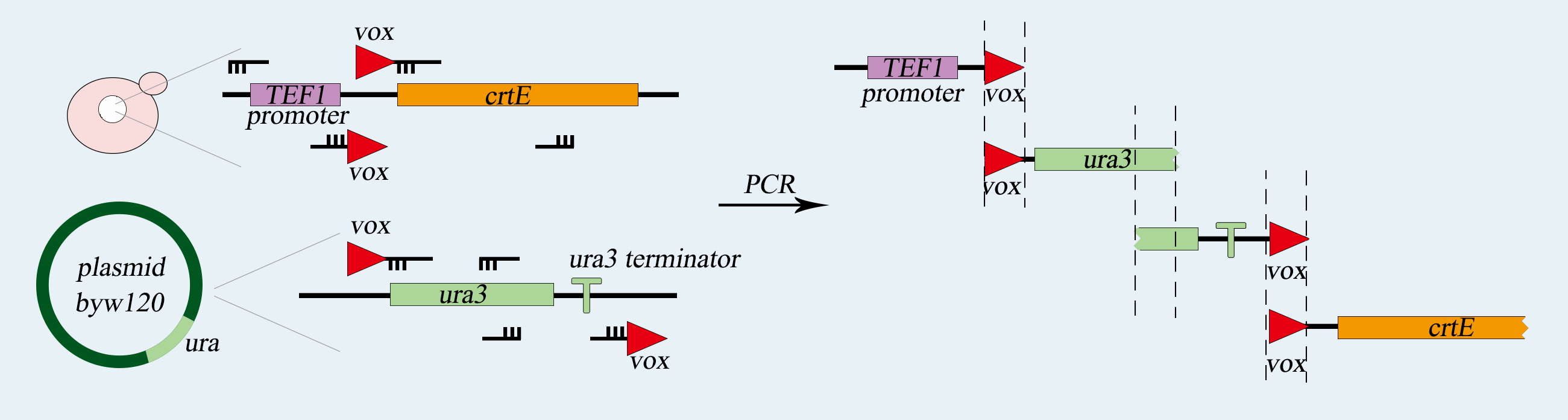
Fig.2-2. The obtention of vox-ura3-terminators-vox structure.
2) Construction of vox-RFP-terminators-vox Structure
This structure has a great similarity to the vox-ura3-terminator-vox structure above. Therefore, it is easy to construct because we only need to change the ura3 gene to the yEmRFP gene. We use PCR to amplify five basic prats including TEF promoter, yEmRFP gene, Adh1 terminator, ura3-terminator and β-carotene gene. Among them, yEmRFP gene and ura3-terminator are flanked by vox locus. Then we use overlap PCR to combine these parts together. After that we use the lithium acetate conversion method to transfer this composite part into TVUVC. We screen for the correctly transformed cell by the 5-FOA plate. This part will integrate into SynⅤ by homologous recombination, and we will get another desired strain called TVRVC.

Fig.2-3. The obtention of vox-RFP-terminators-vox structure.

Fig.2-3. The obtention of vox-RFP-terminators-vox structure.
3) Verification of yEmRFP in the PVRVC
The verification of yEmRFP is carried out by using colony PCR to amplify the homologous arm-TEF promoter-vox-RFP gene and terminator-vox-crtE gene, which determines the existence of vox sites and yEmRFP gene. Then we can detect the red fluorescence.
4) Method of Red Fluorescence Assay
We use a variant of the mCherry red fluorescent protein (RFP). The variant sequence was codon-optimized for the expression in Saccharomyces cerevisiae as yeast-enhanced mRFP (yEmRFP) and can combine fluorescence and a purple visible phenotype. Unfortunately, the RFP can’t be directly observed by bare eyes, we decided to use the Fluorescence spectrophotometer(F-2500) and use OD600 to determination cell concentration. Meanwhile, we will observe using Fluorescence microscopy for fluorescent proteins expression. The red color can be observed if yEmRFP is expressed.
5) Construction of Vika System
We use a common expression vector plasmid, pRS416, to load Vika part. First of all, we use corresponding restriction endonuclease Sal1 and Not1 to cut plasmid pRS416 and plasmid pRS415-Vika, a gift from Y.J lab, and then use T4 DNA ligase to link them together, we can obtain the complete device we want. Finally, we transform this device into BY4742 by the lithium acetate conversion method, and we screen for the correctly transformed cell by using the Sc-Ura plate. BY4742 is a haploid with mating type of α.
We also use another plasmid pRS413 with previous method.
6) The Induction of Mating
The Saccharomyces cerevisiae called PVRVC is haploid with mating type of a. At first, we cultivate pRS416-Vika in Sc-Ura medium without glucose for three hours. To induce the expression of Vika, they will be cultured to saturation in Sc medium with raffinose and galactose for twelve hours. After that, recombinase Vika are induced to express and we make α-pRS416-Vika cell and a-TVRVC cell mate in YPD medium for eight hours. Two types cells are fused and form diploid yeasts, in which recombinase Vika bind with vox locus, and then delete yEmRFP gene and Adh1 terminator flanked by vox sites. After the Mating Switcher, β-carotene expresses and the color of cell will transform from white to orange. At last we smear yeast solution on Sc-Ura-Leu plate to screen for the correctly mating cell. We can judge the existence of recombinase Vika by the color of the colony, and obtain the efficiency of mating.
7) Culture and Expression Condition of Saccharomyces cerevisiae in this Experiment
Traditional YPD culture medium (22g/L glucose, 20g/L peptone, 10g/L yeast extracts) is used by us. Sc-Ura solid culture medium (22g/L glucose, 6.7g/L yeast nitrogen base, 1.224g/L nutrient deficiency mixture without Ura, His, Leu and Trp, 20g/L agar powder, 5mg/L Trp, His and Leu) is used to screen for correctly transformed cell. 5-FOA solid culture medium (22g/L glucose, 6.7g/L yeast nitrogen base, 1.224g/L dropout, 20g/L agar powder, 1ml/L His, Trip, Leu and 2.5ml/L Ura) is used to screen for correctly transformed cell. Sc medium with raffinose and galactose culture medium (20g/L raffinose, 6.7g/L yeast nitrogen base, 1.224g/L nutrient deficiency mixture without Ura, His, Leu and Trp, 20g/L agar powder, 10x galactose, 5mg/L Trp, His and Leu) is used to induce to express vika recombinase. Sc-Ura-Leu solid culture medium(22g/L glucose, 6.7g/L yeast nitrogen base, 1.224g/L nutrient deficiency mixture without Ura, His, Leu and Trp, 20g/L agar powder, 5mg/L Trp and His) is used to screen for the correctly mating cell. All the cells are cultured in 5mL medium at 30℃ with shaking speed of 220rpm.
EXPECTED RESULTS
In our design, Mating Switcher is a means of gene regulation. We can transform from one functional system to another system through this switch conveniently. To show the function of Mating Switcher more intuitively, we construct this RFP system to be a characterization.
First, we can successfully construct the corresponding expression vector.
Second, we expect to observe the expression of yEmRFP before the switch is on. We expect red fluorescent protein to be observed under a fluorescence microscope, and it is desirable that the fluorescence spectrophotometer can measure a high and stable fluorescence value.
Third, Vika-vox system can play a role on function conversion. It is means that β-carotene gene can express after mating. We will observe orange colonies on nutrient label medium.
REFERENCE
[1] Lin, Qiuhui et al. “Robust Orthogonal Recombination System for Versatile Genomic Elements Rearrangement in Yeast Saccharomyces Cerevisiae.” Scientific Reports 5 (2015).
[2] Karimova, Madina et al. “Vika/vox, a Novel Efficient and Specific Cre/loxP-like Site-Specific Recombination System.” Nucleic Acids Research 41.2 (2013).
[3]Verwaal, René et al. “High-Level Production of Beta-Carotene in Saccharomyces Cerevisiae by Successive Transformation with Carotenogenic Genes from X.anthophyllomyces Dendrorhous .” Applied and Environmental Microbiology 73.13 (2007): 4342–4350. PMC. Web. 27 Oct. 2017.
Show less