Erwan Eriau (Talk | contribs) m (grammar) |
Erwan Eriau (Talk | contribs) m |
||
| Line 20: | Line 20: | ||
Here we wanted to answer a single question: '''As p5 recognized M13ori, will the bacterium produces only PLPs or will it also produces replicative phages ?'''. | Here we wanted to answer a single question: '''As p5 recognized M13ori, will the bacterium produces only PLPs or will it also produces replicative phages ?'''. | ||
| + | |||
| + | == Introduction == | ||
| + | |||
| + | Modelling is an essential aspect of engineering, and we wanted to engineer a novel specific phage targeted to Xylella fastidiosa. Apart from a preliminary exercise model used to test our understanding of our system, most of our work was done using a (heavily) adapted version of an already published model of M13 physiology. Modelling allows engineers to explore several avenues of research very quickly and to discard those less likely to succeed early on and at all stages of prototyping and development. For this back and forth betwen wet- and dry-lab to be meaningfull, it needs to be validated against preliminary data, which we did by ensuring our model fitted experimental particle profiles obtained by the wetlab team. We then investigated wether levels of certain key proteins changed particle composition, before determining the ideal number of plasmid copies to optimize our production for the wetlab team. | ||
==First iteration of the model== | ==First iteration of the model== | ||
| − | We started with an excessively simple model to see whether our understanding of our system was profound enough to allow room for such simplifications. We made the following assumptions in this spirit: | + | We started with an excessively simple model to see whether our understanding of our system was profound enough to allow room for such simplifications. This was also the occasion for us to "cut our teeth" modelling, Igem being an occasion for us to expand our initially limited skills. |
| + | We made the following assumptions in this spirit: | ||
Which led to the following model: | Which led to the following model: | ||
Revision as of 02:30, 30 October 2017
Engineered M13 modelling
Contents
With our project, KILL XYL we set to engineer a cure against the disease caused by the plant pathogen Xylella fastidiosa. One of our approaches is to engineer phage-like particles (PLPs), that will deliver toxic genes into the bacterium. Phage systems, like M13, have been employed in biotechnological applications, most prominently in the identification and maturation of medically-relevant binding molecules through phage display [1].
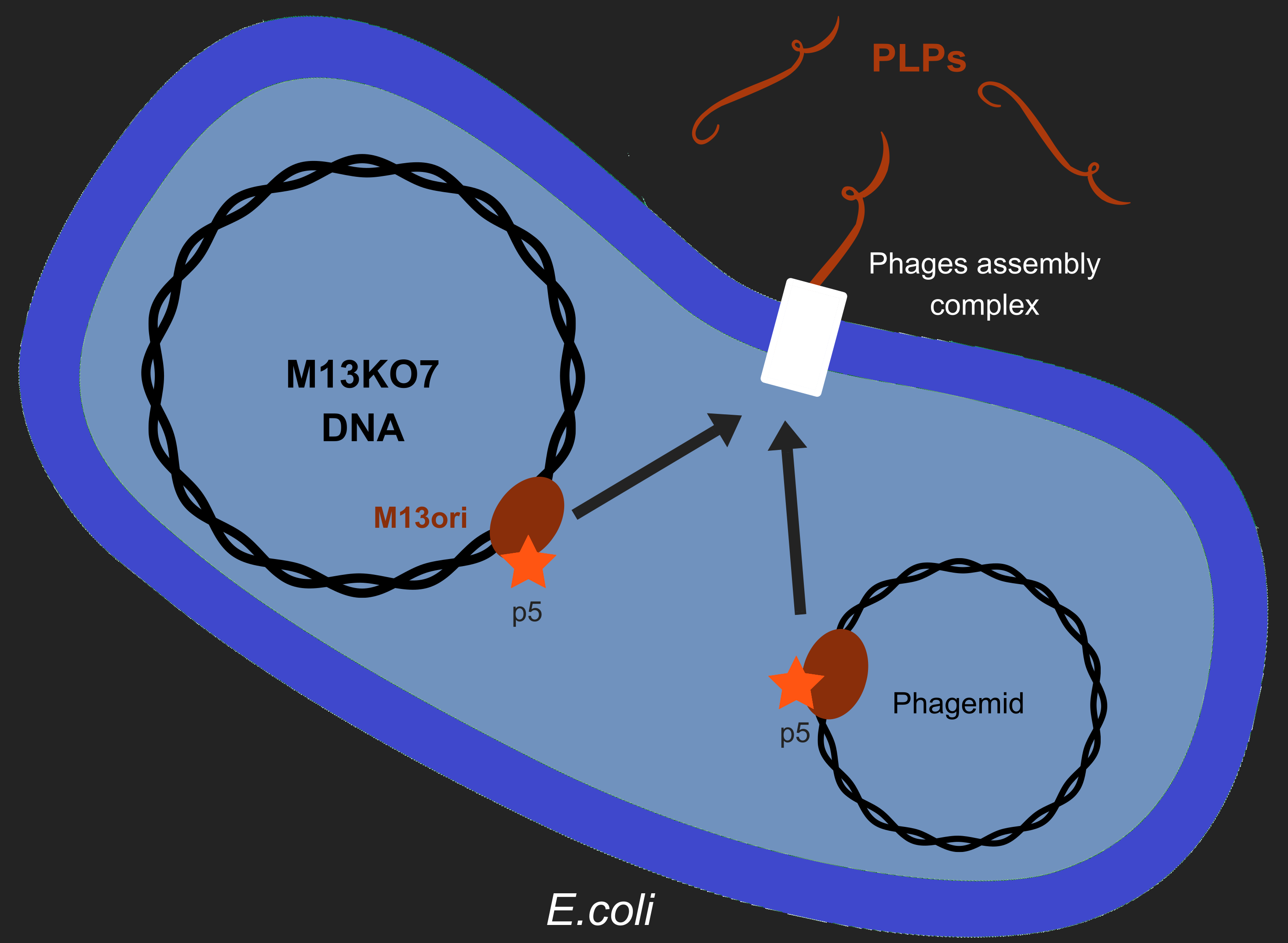
However, production of phages such as M13 has longterm consequences in E. coli, especially regarding metabolism[2]. M13 has a particular life-cycle among phages: it is neither a virulent nor a temperate phage but rather engages in chronic infection. One of the crucial events of it's multiplication is the packaging of its own DNA in the capsid. To do so, M13's protein 5 (p5) targets M13 origin of replication (M13ori) and packages the single-strand DNA. This step is crucial to initiate formation of the phage particle. Afterwards, an assembly complex (made of proteins p1, p4 and p11) assembles the capsid (made of proteins p3, p6, p7, p8, p9) around the ssDNA.
One of the major reasons we chose to use phage-like particles (PLPs) was for our device to be non replicative, so as to prevent the spread of GMOs in the environnement. We designed these PLPs by modifying M13's genome to prevent replication and the addition of a phagemid in the factory cell E.coli. Therefore, to create PLPs we use a modified M13 genome (M13KO7) that has several genes inserts in it M13ori and we integrate a phagemid with a M13ori.
Thanks to Jacques VAN HELDEN interview we put attention to the risk of recombination between M13KO7 and the phagemid. This question matter because we aim to create PLPs that aren't able to replicate and thus spread in the environment. Therefore, we test this possibility along our wetlab tests.
Here we wanted to answer a single question: As p5 recognized M13ori, will the bacterium produces only PLPs or will it also produces replicative phages ?.
Introduction
Modelling is an essential aspect of engineering, and we wanted to engineer a novel specific phage targeted to Xylella fastidiosa. Apart from a preliminary exercise model used to test our understanding of our system, most of our work was done using a (heavily) adapted version of an already published model of M13 physiology. Modelling allows engineers to explore several avenues of research very quickly and to discard those less likely to succeed early on and at all stages of prototyping and development. For this back and forth betwen wet- and dry-lab to be meaningfull, it needs to be validated against preliminary data, which we did by ensuring our model fitted experimental particle profiles obtained by the wetlab team. We then investigated wether levels of certain key proteins changed particle composition, before determining the ideal number of plasmid copies to optimize our production for the wetlab team.
First iteration of the model
We started with an excessively simple model to see whether our understanding of our system was profound enough to allow room for such simplifications. This was also the occasion for us to "cut our teeth" modelling, Igem being an occasion for us to expand our initially limited skills. We made the following assumptions in this spirit:
Which led to the following model:
| Assumption | Reasonning |
|---|---|
| Neglecting cooperative effects | |
| Constant rate of p5 transcription/traduction | |
| One M13KO7 per factory cell | |
| Phage assembly isn't a limiting factor | |
| Neglecting stochasticity |
Second iteration
This time, we decided to adopt the model of a "wild-type" M13's biology [2][3] to fit our own engineered version of M13 and better inform wet lab experiments, as well as explain some of their preliminary findings!
Model validation
compare particle profile to that obtained in wet lab
--> Insufficient p3
could cause the packing of several DNA molecules per phage.
What is the ideal number of plasmid copies
Wet-lab and dry-lab back and forth
References
- ↑ Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).
- ↑ 2.0 2.1 Smeal et al, Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017
- ↑ Smeal et al, Simulation of the M13 life cycle II: Investigation of the control mechanisms of M13 infection and establishment of the carrier state, Virology, 500, January 2017