| Line 278: | Line 278: | ||
<h2><i class="fa fa-university" aria-hidden="true"></i> Overview</h2> | <h2><i class="fa fa-university" aria-hidden="true"></i> Overview</h2> | ||
| − | <p>The models we built included five parts. First, We established a bistable model to analyze the feasibility of functional switch of cells. | + | <p>The models we built included five parts. First, We established a bistable model to analyze the feasibility of functional switch of cells. Then, We used software to model and simulate the functional switch process after cell mating. And we built a RFP expression model to simulate the expression RFP in yeast cells with CUP1 promoter. What’s more, we constructed a model of adsorption to simulate the combination of heavy metal ions and heavy metal-treated proteins inside the yeast cells. Finally, we worked with teammates of human practice to draw the World Copper and Cadmium Pollution Map and established a preliminary World Copper and Cadmium Pollution Database.</p> |
</div> | </div> | ||
| Line 303: | Line 303: | ||
</div> | </div> | ||
<div class="collapse-card__body"> | <div class="collapse-card__body"> | ||
| − | <p>Bistability is a common phenomenon in single-cell microbes, | + | <p>Bistability is a common phenomenon in single-cell microbes, where two types of cell phenotype coexist. Bistability is very important for many single-cell microbes adapting to environmental changes. Single-cell microbes can choose the appropriate form according to changes of the environment, and bistability is the basis for achieving this change.</p> |
| − | <p> | + | <p>The reason for the existence of bistability in single-cell microbes is complex, and it is generally thought to be related to the positive feedback of the gene network. We simulate the bistability in single-celled microbes by establishing a simplified gene regulation model.</p> |
| Line 325: | Line 325: | ||
</p> | </p> | ||
| − | <p>Note that all of these formulations of | + | <p>Note that all of these formulations of derivation of Hill function from mass action kinetics |
assume that the protein has n sites to which ligands can bind. In practice, however, the Hill Coefficient n rarely provides an accurate approximation of the number of ligand binding sites on a protein.[2] We assume that the Hill Coefficient is 2 according to experience. | assume that the protein has n sites to which ligands can bind. In practice, however, the Hill Coefficient n rarely provides an accurate approximation of the number of ligand binding sites on a protein.[2] We assume that the Hill Coefficient is 2 according to experience. | ||
</p> | </p> | ||
| Line 355: | Line 355: | ||
\] | \] | ||
</p> | </p> | ||
| − | <p>Draw the parameters equation, can be obtained under the range of parameters in bistability:</p> | + | <p>Draw the parameters equation, we can be obtained under the range of parameters in bistability:</p> |
<img 6> | <img 6> | ||
| Line 463: | Line 463: | ||
| − | <p>Modeling the functional switch function of yeast cells after mating. After the yeast cells | + | <p>Modeling the functional switch function of yeast cells after mating. After the yeast cells mating, the expression of the functional gene(RCu) is activated, thereby inhibiting the expression of the gene(Cu), and the expression of the downstream gene(Cr) of the inhibited gene(Cu) is activated. As the process involves many genes and the regulatory process is complex, we use Cell Illustrator Online 5.0 to simulate the process. |
</p> | </p> | ||
| Line 488: | Line 488: | ||
<hr> | <hr> | ||
| − | <p>The rate of transcription and translation depends on the concentration of DNA and mRNA, but the transcription and translation process does not directly consume DNA and mRNA, so in these two processes use the auxiliary connections. In addition, we establish the self-degradation process of mRNA and Cu protein. | + | <p>The rate of transcription and translation depends on the concentration of DNA and mRNA, but the transcription and translation process does not directly consume DNA and mRNA, so in these two processes we use the auxiliary connections. In addition, we establish the self-degradation process of mRNA and Cu protein. |
</p> | </p> | ||
<img> | <img> | ||
| Line 522: | Line 522: | ||
| − | <p>We add the substance youdao to our model to simulate the expression of the gene RCu. | + | <p>We add the substance youdao to our model to simulate the expression of the gene RCu. It has two advantages: youdao's self-degradation process makes the inhibitor gradually self-degraded over time, so you can observe whether the effect of RCu protein on Cu DNA is irreversible; youdao can simulate the influence of the addition of foreign substance to yeast, so as to increase the scope of using of our model.</p> |
| − | <p>The rate of transcription and translation depends on the concentration of RCu DNA and RCu mRNA, but the transcription and translation process does not directly consume DNA and mRNA, so in these two processes use the auxiliary connections. In addition, we establish the self-degradation process of RCu mRNA and RCu protein.</p> | + | <p>The rate of transcription and translation depends on the concentration of RCu DNA and RCu mRNA, but the transcription and translation process does not directly consume DNA and mRNA, so in these two processes we use the auxiliary connections. In addition, we establish the self-degradation process of RCu mRNA and RCu protein.</p> |
<h4>Modeling of the binding of protein(RCu) and DNA(Cu)</h4> | <h4>Modeling of the binding of protein(RCu) and DNA(Cu)</h4> | ||
| Line 605: | Line 605: | ||
<div class="collapse-card__body"> | <div class="collapse-card__body"> | ||
<h4>Overview</h4> | <h4>Overview</h4> | ||
| − | <p> In our experiment we use engineered yeast cells to absorb and enrich heavy metals such as cooper and cadmium. At first heavy metal ions diffuse into the cell surface from the liquid phase body, and then heavy metal ions are combined with those heavy metal-treated proteins inside the yeast cells.</p> | + | <p> In our experiment we use engineered yeast cells to absorb and enrich heavy metals such as cooper and cadmium. At first, heavy metal ions diffuse into the cell surface from the liquid phase body, and then heavy metal ions are combined with those heavy metal-treated proteins inside the yeast cells.</p> |
| Line 621: | Line 621: | ||
<h4>Summary</h4> | <h4>Summary</h4> | ||
| − | <p>Treating heavy metal pollution by means of biosorption is a complicated process. First, it is very meaningful to study the growth of yeast in heavy metal ions solution. Considering that the toxic effects of heavy metal ions on yeast can’t be ignored, we use the matrix inhibition growth model to simulate the growth kinetics of yeast in heavy metal ions solution. Next, we decide to study the process of biological adsorption from the thermodynamic and kinetic point of view. In terms of thermodynamics, we use the basic thermodynamic function to explain the adsorption process, and the conclusions can guide | + | <p>Treating heavy metal pollution by means of biosorption is a complicated process. First, it is very meaningful to study the growth of yeast in heavy metal ions solution. Considering that the toxic effects of heavy metal ions on yeast can’t be ignored, we use the matrix inhibition growth model to simulate the growth kinetics of yeast in heavy metal ions solution. Next, we decide to study the process of biological adsorption from the thermodynamic and kinetic point of view. In terms of thermodynamics, we use the basic thermodynamic function to explain the adsorption process, and the conclusions can guide further optimization of the biosorption in addition, different static adsorption models are used to simulate the adsorption process, and the conclusions are able to explain the mechanism of the part of the biosorption process. Then we discuss the change of heavy metal ions with time in the process of biosorption from the point of view of dynamics, and compare with the actual measured data.</p> |
<h4>Yeast growth model</h4> | <h4>Yeast growth model</h4> | ||
| − | <p>Heavy metal ions inhibit the growth of yeast. In order to describe the kinetics of cell growth accurately,these | + | <p>Heavy metal ions inhibit the growth of yeast. In order to describe the kinetics of cell growth accurately,these factors should be taken into account. Unlike the traditional Monod equation, Andrew equation takes the presence of matrix anticompetitive inhibition into consideration.</p> |
<p> | <p> | ||
| Line 753: | Line 753: | ||
<p>\[\frac{{[MX]}}{{{M_e}}} = K({X_0} - [MX])\]</p> | <p>\[\frac{{[MX]}}{{{M_e}}} = K({X_0} - [MX])\]</p> | ||
<h4> Biomass adsorption kinetics</h4> | <h4> Biomass adsorption kinetics</h4> | ||
| − | <p>The biosorption process can be divided into two stages. The first stage occurs on the cell wall surface, and | + | <p>The biosorption process can be divided into two stages. The first stage occurs on the cell wall surface, and is mainly about the physical adsorption and ion exchange process which is going very fast. The second stage, also known as active adsorption,is mainly about chemical adsorption, and metal ions at this stage can be transported through the active into the cell. This stage consumes the energy generated by cell metabolism, which is carried out very slowly.<br> |
Puranik and Paknikar describe the adsorption kinetics quantitatively in mathematical models. This mathematical model is based on the following two assumptions: | Puranik and Paknikar describe the adsorption kinetics quantitatively in mathematical models. This mathematical model is based on the following two assumptions: | ||
</p> | </p> | ||
| Line 768: | Line 768: | ||
\({K_t}\) : Rate constant. | \({K_t}\) : Rate constant. | ||
</p> | </p> | ||
| − | + | <img src=""> | |
| − | + | <p>Fig.14.</p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | "> | + | |
| − | <p | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<p>According to the expression of θ (t), the physical meaning of 1 - θ (t) is that the remaining adsorption sites account for the percentage of total adsorption active sites at time t. <br> | <p>According to the expression of θ (t), the physical meaning of 1 - θ (t) is that the remaining adsorption sites account for the percentage of total adsorption active sites at time t. <br> | ||
Integral: | Integral: | ||
| Line 831: | Line 806: | ||
<h3 class="collapse-card__title"> | <h3 class="collapse-card__title"> | ||
<i class="fa fa-random"></i> | <i class="fa fa-random"></i> | ||
| − | Construction of | + | Construction of RFP expression Model |
</h3> | </h3> | ||
</div> | </div> | ||
| Line 837: | Line 812: | ||
<h4>Overview</h4> | <h4>Overview</h4> | ||
| − | <p>Our goal is to simulate the RFP expression of the yeast cell with | + | <p>Our goal is to simulate the RFP expression of the yeast cell with CUP1 promoter and compare with the wild type yeast which has only a normal promoter and RFP gene. |
With this goal, the first thing we needed to do is using computational method simulate the biological process and figure out whether our design is feasible. | With this goal, the first thing we needed to do is using computational method simulate the biological process and figure out whether our design is feasible. | ||
</p> | </p> | ||
| Line 859: | Line 834: | ||
\([aACE1]\)is the concentration of active transcription factor ACE1;<br> | \([aACE1]\)is the concentration of active transcription factor ACE1;<br> | ||
\({k_1}\)is reaction rate constant.<br> | \({k_1}\)is reaction rate constant.<br> | ||
| − | If it is higher than a threshold value | + | If it is higher than a threshold value, the concentration of active transcription factor will get decreased.<br> |
\[[aACE1] = {k_{\rm{d}}} \times [ACE1] \times {[C{u^{2 + }}]^{{\rm{ - }}1}}\]<br> | \[[aACE1] = {k_{\rm{d}}} \times [ACE1] \times {[C{u^{2 + }}]^{{\rm{ - }}1}}\]<br> | ||
\({k_{\rm{d}}}\) is the restrain constant; | \({k_{\rm{d}}}\) is the restrain constant; | ||
| − | The process of combination of transcription factor and promoter and RNA polymerase | + | The process of combination of transcription factor and promoter and RNA polymerase which drives mRNA transcription can be described as following equation :<br> |
\[{\rm{dNTP + DNA + E}}{\leftrightarrow }{\rm{ES}}{\leftrightarrow }DNA + mRNA + E\] <br> | \[{\rm{dNTP + DNA + E}}{\leftrightarrow }{\rm{ES}}{\leftrightarrow }DNA + mRNA + E\] <br> | ||
E is the RNA polymerase;<br> | E is the RNA polymerase;<br> | ||
K2 is the reaction rate constant of combination ;<br> | K2 is the reaction rate constant of combination ;<br> | ||
K3 is the reaction rate constant of transcription; <br> | K3 is the reaction rate constant of transcription; <br> | ||
| − | The process can be seen as | + | The process can be seen as the kinetics of an enzymatic reaction in Michaelis–Menten kinetics model. The reaction rate can be described as :<br> |
\[{{\rm{V}}_{}}{\rm{ = }}\frac{{{{\rm{V}}_{\max }}[aACE1]}}{{{k_m} + [aACE1]}}\]<br> | \[{{\rm{V}}_{}}{\rm{ = }}\frac{{{{\rm{V}}_{\max }}[aACE1]}}{{{k_m} + [aACE1]}}\]<br> | ||
\({{\rm{V}}_{\max }}\)is the maximum reaction rate;<br> | \({{\rm{V}}_{\max }}\)is the maximum reaction rate;<br> | ||
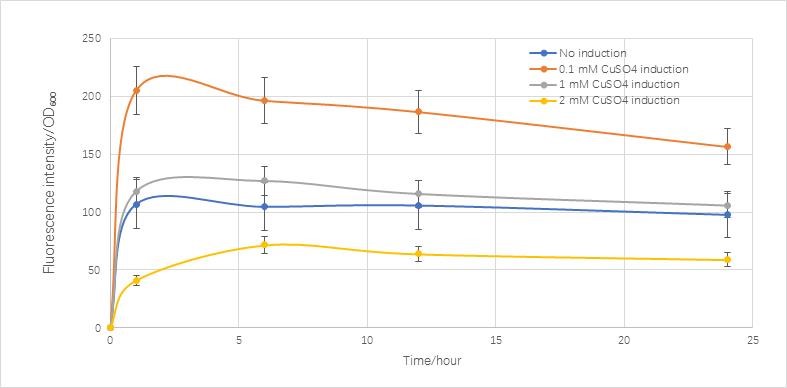
| Line 940: | Line 915: | ||
Compared with the real condition we measured,it is very similar to the real condition except in the final stage the real condition concentration is going to decrease.</p> | Compared with the real condition we measured,it is very similar to the real condition except in the final stage the real condition concentration is going to decrease.</p> | ||
<h4>SUMMARY</h4> | <h4>SUMMARY</h4> | ||
| − | <p>We construct the model of RFP expression in the single cell stage with molecular analysis. The result is similar with the real condition. This model can help us to confirm the CUP1 promoter is effective to be induced by copper ions. It also can help us to confine the best concentration of copper ions to induce and | + | <p>We construct the model of RFP expression in the single cell stage with molecular analysis. The result is similar with the real condition. This model can help us to confirm that the CUP1 promoter is effective to be induced by copper ions. It also can help us to confine the best concentration of copper ions to induce and the maximum expression quantity in a certain copper ions concentration. |
</p> | </p> | ||
| Line 960: | Line 935: | ||
</div> | </div> | ||
<div class="collapse-card__body"> | <div class="collapse-card__body"> | ||
| − | <p>This part is not common in modeling because this part is not a model of our biological system to simulate and predict its behavior. What we do in this part is an assessment of its value and scope of use of our biological systems. In addition, we get a preliminary database of copper and cadmium pollution in the world through the data collection, which is also within the scope of modeling.In this part, we work with teammates who responsible for human practice to carry out the main work of data collection and map processing. Because different countries and regions have different monitoring and treatment methods for copper and cadmium pollution, and many countries and regions even lack monitoring and treatment methods of copper and cadmium pollution, we spent a lot of time to collect data | + | <p>This part is not common in modeling, because this part is not a model of our biological system to simulate and predict its behavior. What we do in this part is an assessment of its value and scope of use of our biological systems. In addition, we get a preliminary database of copper and cadmium pollution in the world through the data collection, which is also within the scope of modeling. In this part, we work with teammates who are responsible for human practice to carry out the main work of data collection and map processing. Because different countries and regions have different monitoring and treatment methods for copper and cadmium pollution, and many countries and regions even lack of monitoring and treatment methods of copper and cadmium pollution, we spent a lot of time to collect data. Even so, we did not collect enough data to conduct detailed data analysis of copper and cadmium pollution worldwide. Nevertheless, our existing work can still lead to some preliminary and regional results for the pollution of copper and cadmium.</p> |
<h4>Data collection</h4> | <h4>Data collection</h4> | ||
| − | <p>As the United States for the detection and processing of pollution is | + | <p>As the United States for the detection and processing of pollution is better, we firstly start from the United States to collect data. We find the National Aquatic Resource Surveys in the EPA (United States Environmental Protection Agency) website. There are four national assessments in NARS: National Coastal Condition Assessment (NCCA), National Lakes Assessment (NLA), National Rivers and Streams Assessment (NRSA), National Wetland Condition Assessment (NWCA). Only the NWCA has the data of copper and cadmium, so we use it as one of the data sources of our maps. The methods and standards used in NWCA are shown in EPA websites.</p> |
<p>The methods we used to collect European data is similar to that of the United States. We use the FOREGS-EuroGeoSurveys Geochemical Baseline Database as one of data sources of our maps. The methods and standards used are shown in FOREGS-EuroGeoSurveys Geochemical Baseline Database websites.<p> | <p>The methods we used to collect European data is similar to that of the United States. We use the FOREGS-EuroGeoSurveys Geochemical Baseline Database as one of data sources of our maps. The methods and standards used are shown in FOREGS-EuroGeoSurveys Geochemical Baseline Database websites.<p> | ||
<p>The data collection process of China is more difficult than that of Europe and the USA, although China also has a comprehensive monitoring and treatment system for copper and cadmium pollution, these data have not published in detail, so we cannot get these authoritative national census data.</p> | <p>The data collection process of China is more difficult than that of Europe and the USA, although China also has a comprehensive monitoring and treatment system for copper and cadmium pollution, these data have not published in detail, so we cannot get these authoritative national census data.</p> | ||
| − | <p>So we turn to other sources to collect data | + | <p>So we turn to other sources to collect data of China. The team of Jianchao Li from Shanxi Normal University retrieved 2450 papers on six kinds of heavy metal pollution (Cd, Pb, Zn, Zn, Cu, Cu, Cr) in China's soil for nearly ten years. After finishing the analysis, the data of 850 papers has plotted the distribution of heavy metal pollution, and provided a detailed distribution of the key pollution areas. The work was published in June 2016 at Bulletin of Environmental Contamination and Toxicology.</p> |
<p><i>Duan Q., Lee J., Liu Y., Chen H., Hu H., (2016) Distribution of Heavy Metal Pollution in Surface Soil Samples in China: A Graphical Review. Bull Environ Contam Toxicol. Jun 24.</i></p> | <p><i>Duan Q., Lee J., Liu Y., Chen H., Hu H., (2016) Distribution of Heavy Metal Pollution in Surface Soil Samples in China: A Graphical Review. Bull Environ Contam Toxicol. Jun 24.</i></p> | ||
| − | <p>For the data of other regions, we did not find similar data sources, so we refer to the practice of the Jiaochao Li team, through the web of science core collection on the literature in recent years, we retrieve 309 sets of data | + | <p>For the data of other regions, we did not find similar data sources, so we refer to the practice of the Jiaochao Li team, through the web of science core collection on the literature in recent years, we retrieve 309 sets of data. Although these data is not enough Detailed global analysis, we draw some preliminary conclusions.</p> |
| Line 979: | Line 954: | ||
<h4>Map Drawing</h4> | <h4>Map Drawing</h4> | ||
| − | <p>We stored data we collected as .mat file in MATLAB, and then we use the | + | <p>We stored data we collected as .mat file in MATLAB, and then we use the worldmap and our map drawing command to draw our Global Cu and Cd Pollution Map.</p> |
| Line 1,010: | Line 985: | ||
<div class="centered-y"> | <div class="centered-y"> | ||
<h2>Brazil</h2> | <h2>Brazil</h2> | ||
| − | <p>We get data from web of science, so it cannot represent Brazilian soil copper concentration comprehensively. Seen form the data got from web of science, many regions in Brazil have a high and even | + | <p>We get data from web of science, so it cannot represent Brazilian soil copper concentration comprehensively. Seen form the data got from web of science, many regions in Brazil have a high and even extremely high concentration.</p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 1,055: | Line 1,030: | ||
<p>From the pollution map we can see the following:<br> | <p>From the pollution map we can see the following:<br> | ||
| − | 1, The pollution in Asia, Africa and Latin America is more serious in the United | + | 1,The pollution in Asia, Africa and Latin America is more serious in the United |
States and Europe: Although we don’t have comprehensive data for Africa and Latin | States and Europe: Although we don’t have comprehensive data for Africa and Latin | ||
Revision as of 12:46, 31 October 2017
/* OVERRIDE IGEM SETTINGS */
Model
Overview
The models we built included five parts. First, We established a bistable model to analyze the feasibility of functional switch of cells. Then, We used software to model and simulate the functional switch process after cell mating. And we built a RFP expression model to simulate the expression RFP in yeast cells with CUP1 promoter. What’s more, we constructed a model of adsorption to simulate the combination of heavy metal ions and heavy metal-treated proteins inside the yeast cells. Finally, we worked with teammates of human practice to draw the World Copper and Cadmium Pollution Map and established a preliminary World Copper and Cadmium Pollution Database.