Cccxyyyyyyyy (Talk | contribs) |
|||
| Line 399: | Line 399: | ||
<hr> | <hr> | ||
<h5>1) Construction of <i>vox-ura3-terminator-vox</i> Structure </h5> | <h5>1) Construction of <i>vox-ura3-terminator-vox</i> Structure </h5> | ||
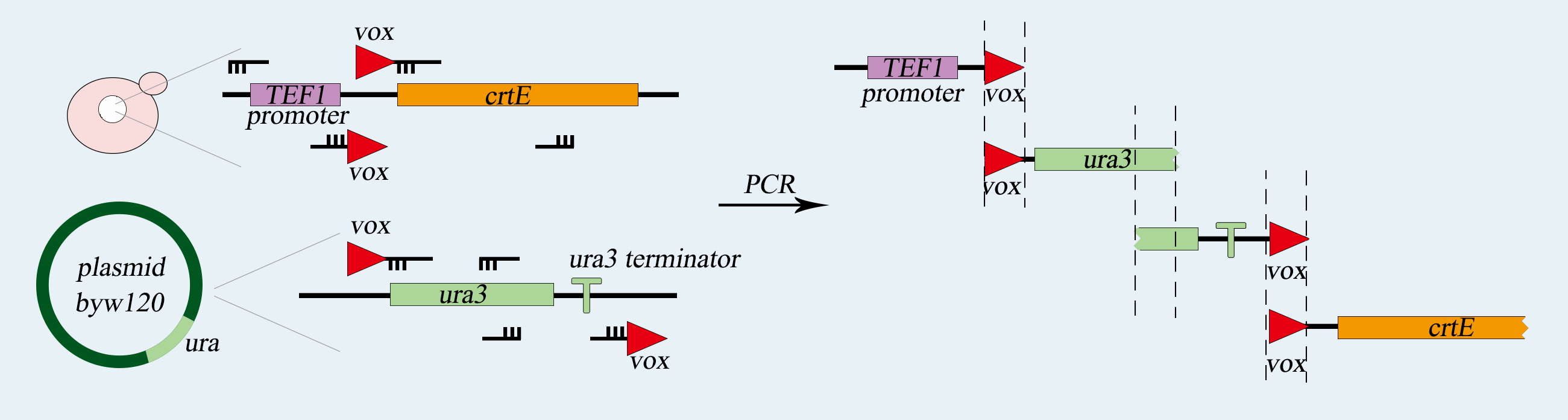
| − | <p> We use <i>synthetic chromosome Ⅴ</i> of <i>Saccharomyces cerevisiae</i> to load our device, which is a single-celled organism called a. First of all, we use PCR to amplify basic parts including <i>TEF</i> promoter, <i>ura3</i> gene, <i>ura3-terminator</i> and <i>β-carotene</i> gene. Among them, <i>ura3</i> gene and <i>ura3-terminator</i> are flanked by <i>vox</i> locus. Then we use overlap PCR to combine these parts together. The next step is transform this composite part into <i>Saccharomyces cerevisiae</i>. We screen for the correctly transformed cell by using the <i>Sc-Ura</i> plate. For the purpose of verifying desired strain <i>PVUVC</i>, we use colony PCR to amplify the <i>TEF promoter-vox-ura3</i> structure and <i>ura3 terminator-vox-β-carotene</i> structure. The length of the strip was observed by agarose gel electrophoresis. | + | <p> We use <i>synthetic chromosome Ⅴ</i> of <i>Saccharomyces cerevisiae</i> to load our device, which is a single-celled organism called a. First of all, we use PCR to amplify basic parts including <i>TEF</i> promoter, <i>ura3</i> gene, <i>ura3-terminator</i> and <i>β-carotene</i> gene. Among them, <i>ura3</i> gene and <i>ura3-terminator</i> are flanked by <i>vox</i> locus. Then we use overlap PCR to combine these parts together. The next step is transform this composite part into <i>Saccharomyces cerevisiae</i>. We screen for the correctly transformed cell by using the <i>Sc-Ura</i> plate. For the purpose of verifying desired strain <i><b>PVUVC</b></i>, we use colony PCR to amplify the <i>TEF promoter-vox-ura3</i> structure and <i>ura3 terminator-vox-β-carotene</i> structure. The length of the strip was observed by agarose gel electrophoresis. |
</p> | </p> | ||
| Line 417: | Line 417: | ||
<h5>2) Construction of <i>vox-RFP-terminators-vox</i> Structure</h5> | <h5>2) Construction of <i>vox-RFP-terminators-vox</i> Structure</h5> | ||
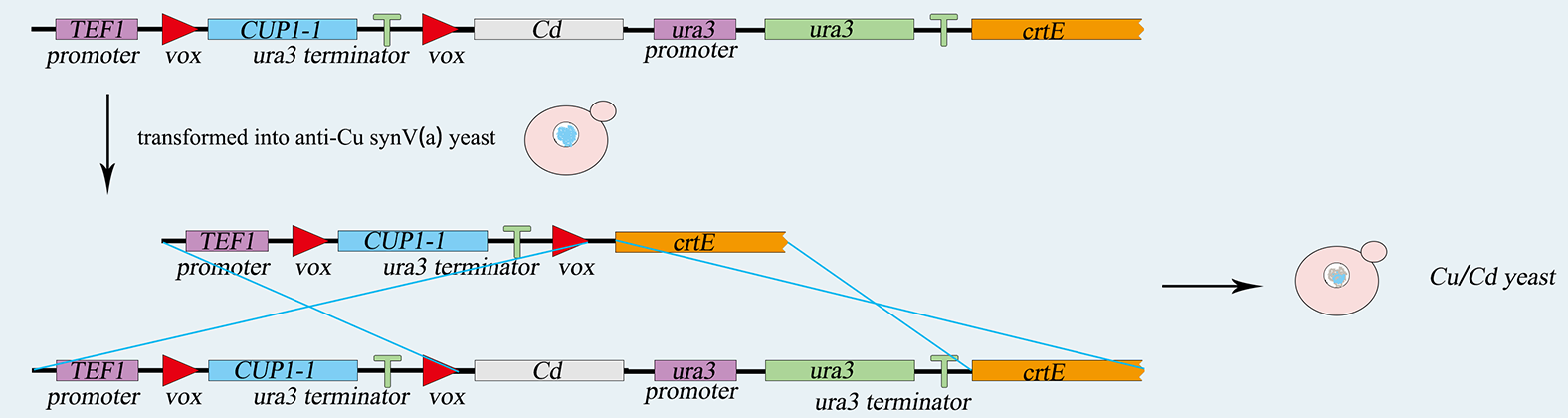
| − | <p> This structure has a great similarity to the <i>vox-ura3-terminator-vox</i> structure above. Therefore, it is easy to construct because we only need to change the <i>ura3</i> gene to the RFP gene. We use PCR to amplify five basic prats including <i>TEF promoter</i>, <i>RFP gene</i>, <i>Adh1 terminator</i>, <i>ura3-terminator</i> and <i>β-carotene</i> gene. Among them, <i>RFP</i> gene and <i>ura3-terminator</i> are flanked by <i>vox</i> locus. Then we use overlap PCR to combine these parts together. After that we use the lithium acetate conversion method to transfer this composite part into <i>TVUVC</i>. We screen for the correctly transformed cell by using the 5-FOA plate. This part will integrate into <i>chromosomeⅤ</i> by homologous recombination, and we will get another desired strain called <i>TVRVC</i>. | + | <p> This structure has a great similarity to the <i>vox-ura3-terminator-vox</i> structure above. Therefore, it is easy to construct because we only need to change the <i>ura3</i> gene to the RFP gene. We use PCR to amplify five basic prats including <i>TEF promoter</i>, <i>RFP gene</i>, <i>Adh1 terminator</i>, <i>ura3-terminator</i> and <i>β-carotene</i> gene. Among them, <i>RFP</i> gene and <i>ura3-terminator</i> are flanked by <i>vox</i> locus. Then we use overlap PCR to combine these parts together. After that we use the lithium acetate conversion method to transfer this composite part into <i><b>TVUVC</b></i>. We screen for the correctly transformed cell by using the 5-FOA plate. This part will integrate into <i>chromosomeⅤ</i> by homologous recombination, and we will get another desired strain called <i><b>TVRVC</b></i>. |
</p> | </p> | ||
| Line 450: | Line 450: | ||
</p> | </p> | ||
<h5>6) The Induction of Mating</h5> | <h5>6) The Induction of Mating</h5> | ||
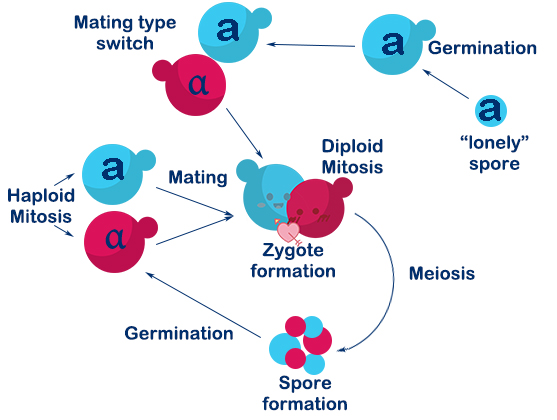
| − | <p> The <i>Saccharomyces cerevisiae</i> called <i>PVRVC</i> is a single-celled organism called a. At first, we cultivate <i>pRS416-vika</i> in <i>Sc-Ura</i> medium without glucose for three hours. To induce the expression of <i>vika</i>, they will culture to saturation in <i>Sc</i> medium with <i>raffinose</i> and <i>galactose</i> for twelve hours. After that, recombinase <i>vika</i> are induced to express and we make <i>α-pRS416-vika</i> cell and a-TVRVC cell mate in <i>YPD</i> medium for eight hours. Two types cells are fused and form diploid yeasts, in which recombinase <i>vika</i> bind with <i>vox</i> locus, and then delete <i>RFP</i> gene and <i>Adh1</i> terminator flanked by <i>vox</i> sites. After the Mating Switcher, <i>β-carotene</i> expresses and the color of cell will transform from white to orange. At last we smear yeast solution on <i>Sc-Ura-Leu</i> plate to screen for the correctly mating cell. We can judge the existence of recombinase <i>vika</i> by the color of the colony, and obtain the efficiency of mating. | + | <p> The <i>Saccharomyces cerevisiae</i> called <i><b>PVRVC</b></i> is a single-celled organism called a. At first, we cultivate <i>pRS416-vika</i> in <i>Sc-Ura</i> medium without glucose for three hours. To induce the expression of <i>vika</i>, they will culture to saturation in <i>Sc</i> medium with <i>raffinose</i> and <i>galactose</i> for twelve hours. After that, recombinase <i>vika</i> are induced to express and we make <i>α-pRS416-vika</i> cell and a-TVRVC cell mate in <i>YPD</i> medium for eight hours. Two types cells are fused and form diploid yeasts, in which recombinase <i>vika</i> bind with <i>vox</i> locus, and then delete <i>RFP</i> gene and <i>Adh1</i> terminator flanked by <i>vox</i> sites. After the Mating Switcher, <i>β-carotene</i> expresses and the color of cell will transform from white to orange. At last we smear yeast solution on <i>Sc-Ura-Leu</i> plate to screen for the correctly mating cell. We can judge the existence of recombinase <i>vika</i> by the color of the colony, and obtain the efficiency of mating. |
</p> | </p> | ||
Revision as of 15:42, 31 October 2017
/* OVERRIDE IGEM SETTINGS */
Design
Background
Human existence on earth is almost impossible without the heavy metals. Even though important to mankind, exposure to them during production, usage and their uncontrolled discharge into the environment has caused lots of hazards to man, other organisms and the environment itself. Heavy metals can enter human tissues and organs via inhalation, diet, and manual handling. As the process of urbanization and industrialization goes deeper and deeper, heavy metal pollution, a noticeable threaten to almost all the creatures, has become an essential problem to solve.
According to our human practice, the situation of heavy metal pollution (copper and cadmium ions) is marked on a world map, and the severity of heavy metal pollution has been increasing all over this map. Places with serious pollution include middle Asia, eastern Asia, southern Europe, and Latin America. In addition, not only fresh water source, but also soil and crops are seriously contaminated by heavy metals. On average, during three out of ten suppers we have, we absorb excess heavy metals over the standard concentration.
Considering the rigorous situation we face, our team decided to design an advanced system for typical toxic heavy metal disposal based on Saccharomyces cerevisiae.