Project Description
Introduction
For years, researchers have used recombinase proteins to genetically modify organisms. However, we noticed that the potential of these proteins in genetic engineering is not fully realised. Scientists can often only use a handful of recombinases within a cell, and they may also cause unwanted side-effects. This limits how experiments can be designed. To address these issues, we have developed the Site-specific, MOdular REcombination (SMORE) toolkit. The biobricks in the SMORE toolkit are all thoroughly tested and documented. They can be expressed in the same cell, which allows for the creation of more complex constructs in synthetic biology.
What is site-specific recombination and why does it matter?
Site-specific recombination (SSR), found mostly in bacteria, viruses and transposons (so-called parasitic DNA elements), is one of the many mechanisms which life utilises to perform genetic recombination [1]. SSR generally consists of a recombinase protein that mediates recombination, and two DNA elements, called target sites, that are similar or identical to each other that the recombinase recognises [2]. Depending on the orientation and the location of the target sites, SSR can perform DNA integration, excision, and exchange. While bacteria use SSR to regulate gene expression and separate two chromosomes during cell division, viruses and transposons use it to mediate chromosomal integration into the bacterial chromosome, hijacking the cellular machinery to replicate themselves. For a more clear picture on how recombination occurs in three-dimensions, see our video below where we extensively explain how SSR occurs, using the popular Cre/Lox system as an example.
Click here for an animated view of how SSR occurs in three dimensions!
With the ability to modify DNA in a precise manner, SSR has been used in various fields of research and industrial applications where genetic engineering is required. Click to see the applications of SSR:
1. Genetic Engineering
a. Conditional gene knockout
Site-specific recombinases are highly efficient in excising DNA segments flanked with target sites; this property has been widely adopted by neuroscientists and developmental biologists to perform conditional gene knockout, where a gene can be deleted in specific time and cell types, and is particularly useful to study genes that are essential for development. For example, Hara et al. showed that autophagy was important not only for adaptation to starvation, but also for preventing degeneration of neurons. They demonstrated it by knocking out Atg5, a protein involved in autophagy, specifically in neurons using Cre/LoxP recombination system, as its complete knockout is lethal to the organism.
Examples:
(1) Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima,
R., Yokoyama, M., Mishima, K., Saito, I., Okano, H., Mizushima, N. (2006) Suppression
of autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441: 885-889
(2) Yang, X., Li, C., Herrera, P., Deng, C. (2002) Generation of Smad4/Dpc4 conditional
knockout mice. Genesis. 32(2): 80-81
b. Transgenesis
SSR has been very useful for inserting genes into designated loci of a genome. This is more advantageous than strategies based on homologous recombination (HR), in that 1) it is more efficient , 2) concatenation effect [2,3] can be eliminated, and 3) selective marker can be removed with ease. Such properties have been exploited into advanced techniques such as recombinase-mediated cassette exchange (RMCE) and serine integrase recombinational assembly (SIRA), where multiple genes can be introduced and exchanged rapidly. They often complement other genetic engineering tools (e.g. CRISPR-Cas system) to perform desired functions.
Examples:
(1) Bischof, J., Maeda, R.A., Hediger, M., Karch, F., Basler, K. (2006)
An optimised transgenesis system for Drosophila using germline-specific φC31 integrases.
Proc Natl Acad Sci. 104(9): 3312-3317
(2) Hammond, A., Galizi, R., Kyrou,K., Simoni,
A., Siniscalchi, C., Katsanos, D., Gribble, M., Baker, D., Marois, E., Russell, S., Burt,
A., Windbichler, N., Crisanti, A., Nolan, T. (2016) A CRISPR-Cas9 gene drive system targeting
female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology.
34(1): 78-83
(3) Colloms, S.D., Merrick, C.A., Olorunniji, F.J., Stark, W.M.,
Smith, M.C.M., Osbourn, A., Keasling, J.D., Rosser, S.J. (2014) Rapid metabolic pathway
assembly and modification using serine integrase site-specific recombination. Nucleic
Acids Res. 42(4): e23
c. Large-scale chromosome modification
Although being gradually replaced by the more recent technology such as CRISPR/Cas system, SSR has been employed to perform large scale DNA modifications (deletion, insertion, and inversion), and is a handy tool for modeling diseases that are caused by chromosome rearrangements. Compared to CRISPR technology, SSR does not rely on the organism’s ability to repair double-stranded breaks (DSBs) by homologous recombination, and is therefore particularly useful in organisms such as bacteria that have limited ability to repair DSBs.
Examples:
(1) Venken, , K.J., He, Y., Hoskins, R.A., Bellen, H.J. (2006) P[acman]:
a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster.
Science. 314(5806): 1747-51
(2) Uemura, M., Niwa, Y., Kakazu, N., Adachi, N., Kinoshita,
K. (2010) Chromosomal manipulation by site-specific recombinases and fluorescent protein-based
vectors. PLoS ONE 5(3): e9846
(3) Enyeart, P.J., Chirieleison, S.M., Dao, M.N., Perutka, J., Quandt, E.M., Yao,
J., Whitt, J.T., Keatinge-Clay, A.T., Lambowitz, A.M., Ellington, A.D. (2013) Generalized
bacterial genome editing using mobile group II introns and Cre-lox. Molecular Systems
Biology. 9(1): 685
2. Research
a. Disease modeling
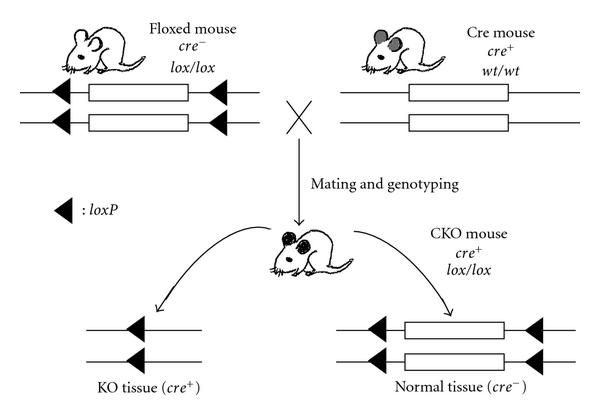
SSR, as an genetic engineering tool, is widely used to simulate and study pathological conditions such as inactivational mutations and chromosomal rearrangements. For example, Barlow et al. used Cre/LoxP mechanism to delete a part of the mouse chromosome 18 to study the 5q- syndrome in human, and they found that the deletion was associated with a change in p53 level, providing insight on its disease mechanism. Figure adapted from Le, Y-Z. (2011) Conditional Gene Targeting: Dissecting the Cellular Mechanisms of Retinal Degenerations. Journal of Opthalmology. Article ID 806783
Examples:
(1) Barlow, J.L., Drynan, L.F., Hewett, D.R., Holmes, L.R., Lorenzo-Abalde,
S., Lane, A.L., Jolin, H.E., Pannell, R., Middleton, A.J., Wong, S.H., Warren, A.J.,
Wainscoat, J.S., Boultwood, J., McKenzie, A.N.J. (2010) A p53-dependent mechanism underlies
macrolytic anemia in a mouse model of human 5q-syndrome. Nature medicine. 16: 59-66
(2) Pan, L., Wang, S., Lu, T., Weng, C., Song, X., Park, J.K., Sun, J., Yang, Z-H.,
Yu, J., Tang, H., McKearin, D.M., Chamovitz, D.A., Ni, J., Xie, T. (2014) Protein competition
switches the function of COP9 from self-renewal to differentiation. Nature. 514(7521):
233-6
b. Cell lineage tracing

Lineage tracing is the tagging of a specific population of cells that allows researchers to study their role in development. This has been very useful in not only the identification of stem cells, but also in the validation of the cancer stem cells theory. Furthermore, Cre/LoxP recombination system has been implemented in the Brainbow system, where more than 100 individual neuron cells can be distinguished from each other in the same image. This unprecedented technique has shone light to the study of connectome, the study of architecture and interaction between neurons in a brain. Figure adapted from Livet et al.
Examples:
(1) Barker, N., van Es, J.H., Kuipers, J., Kujala, P., van den Born,
M., Cozijnsen, M., Haegebarth, A., Korving, J., Begthel, H., Peters, P.J., Clevers, H.
(2007) Identification of stem cells in small intestine and colon by marker gene Lgr5.
Nature. 449: 1003-1007
(2) Livet, J., Weissman, T.A., Kang, H., Draft, R.W., Lu, J., Bennis, R.A., Sanes,
J.R., Lichtman, J.W. (2007) Transgenic strategies for combinatorial expression of fluorescent
proteins in the nervous system. Nature. 450: 56-62
(3) Pei, W., Feyerabend, T.B.,
Rössler, J., Wang, X., Postrach, D., Busch, K., Rode, I., Klapproth, K, Dietlein, N.,
Quadenau, C., Chen, W., Sauer, S., Wolf, S., Höfer, T., Rodewald, H. (2017) Polylox barcoding
reveals haematopoietic stem cell fates realized in vivo. Nature. 548(7668): 456-460
3. Synthetic Biology
a. Complex genetic circuits
Having the ability to perform both irreversible and reversible DNA modification, SSR has been used to create time delay circuits, logic gates, state machines and more. These have found applications on creating complex biosensors -- the detection of specific chemicals using biological pathways. Examples include the detection of multiple gaseous chemicals and integrating the signals using AND, OR, and NOR gates, and the sensitive detection of intracellular miRNA level for developmental biology research.
Examples:
(1) Roquet, N., Soleimany, A.P., Ferris, A.C., Aaronson, S., Lu, T.K.
(2016) Synthetic recombinase-based state machines in living cells. Science. 353(6297):
aad8559
(2) Weinberg, B.H., Hang Pham, N.T., Caraballo, L.D., Lozanoski, T., Engel, A.,
Bhatia, S., Wong, W.W. (2017) Large-scale design of robust genetic circuits with multiple
inputs and outputs for mammalian cells. Nature biotechnology. 35(5): 453-462
(3) Müller, M., Ausländer, S., Spinnler, A., Ausländer, D., Sikorski, J., Folcher,
M., Fussenegger, M. (2017) Designed cell consortia as fragrance-programmable analog-to-digital
converters. Nature chemical biology. 13: 309-316
(4) Lapique, N., Benenson, Y.
(2014) Digital switching in a biosensor circuit via programmable timing of gene availability.
Nature chemical biology. 10: 1020-1027
b. Directed evolution
Evolution is the change in the genetic content of a population of organisms through processes such as mutagenesis and natural selection. The 4.5 billion years of evolution have allowed organisms to diversify and generate numerous traits or combinations of traits; however, evolution itself is a slow process, and sometimes desirable traits may not be easily found in nature. This can sped up by direction evolution, a method to mimic the process of evolution in an artificial way. Although there are many established methods of directed evolution, one of the most extraordinary technologies is SCRaMbLE, which generated synthetic chromosomes in yeast. In SCRaMblE, multiple recombinations sites were included. In the presence of recombinase, the sequences in the synthetic will go through random inversion, deletion, or duplication events. This could potentially be applied not only to screening for the yeast strains with the best performance for a particular application, but also to facilitate research in the minimal genome. It is envisaged that similar system could be applied to various model organisms to facilitate research.

Examples:
(1) Dymond, J.S., Richardson, S.M., Coombes, C.E., Babatz, T., Müller,
H., Annaluru, N., Blake, W.J., Schwerzmann, J.W., Dai, J., Lindstrom, D.L., Boeke, A.C.,
Gottschling, D.E., Chandrasegaran, S., Bader, J.S., Booke, J.D. (2011) Synthetic chromosome
arms function in yeast and generate phenotypic diversity by design. Nature. 477(7365):
471-476
(2) Shen, Y., Stracquadanio, G., Wang, Y., Yang, K., Mitchell, L.A., Xue, Y.,
Cai, Y., Chen, T., Dymond, J.S., Kang, K., Gong, J., Zeng, X., Zhang, Y., Li, Y., Feng,
Q., Xu, X., Wang, J., Wang, J., Yang, H., Booke, J.D., Bader, J.S. (2016) SCRaMbLE generates
designed combinatorial stochastic diversity in synthetic chromosomes. Genome research.
26: 36-49
4. Medical application
a. Diagnostics
Accurate diagnosis plays a decisive role in effective disease treatment. Therefore, a large amount of research effort is put towards its improvement. Particularly, recent years have seen a growing interest in the in vivo diagnostics. Using SSR and synthetic biology, researchers have engineered bacteria that normally inhabit human gut into surveillance machines that sense the gut environment and microbiota. They could potentially be used to record a patient’s past exposure of infections, informing clinicians to prescribe more appropriate therapy.
The iGEM team in ETH Zurich in 2016 has used SSR as a tool to develop E. coli as a surveillance machine to track inflammatory bowel disease (IBD)! Check out their page here: https://2016.igem.org/Team:ETH_Zurich
Examples:
(1) Rubens, J.R., Selvaggio, G., Lu, T.K. (2016) Synthetic mixed-signal
computation in living cells. Nature communications. 7:11658
(2) Mimee, M., Tucker,
A.C., Voigt, C.A., Lu, T.K. (2015) Programming a human commensal bacterium, Bacteroides
thetaiotaomicron, to sense and respond to stimuli in murine gut microbiota. Cell systems.
1:62-71
b. Gene therapy
Gene therapy holds promise in curing diseases previously thought to be impossible to treat. One of the many challenges it faces is the need to lengthen the gene expression time while avoiding its permanent integration into the host genome. A prime example is the use of gene therapy to temporarily stimulate osteogenesis in stem cells, thereby healing bone defects. To enhance the effect of gene therapy, researchers use SSR to circularize the gene fragments transfected into the cells. As circular DNA is less subject to DNA degradation than linear DNA is, this method allows the gene to be expressed for a longer period of time, enhancing the healing potential of the stem cells. Also, SSR is being investigated to cure HIV, and similar diseases such as T-cell leukemia that are caused by retroviruses.

Examples:
(1) Lo, S., Li, K., Chang, Y., Hsu, M., Sung, L., Vu, T. A., Hu, Y. (2017)
Enhanced critical-size calvarial bone healing by ASCs engineered with Cre/loxP-based
hybrid baculovirus. Biomaterial. 124: 1-11
(2) Sarker, I., Hauber, I., Hauber,
J., Buchholz, F. (2007) HIV-1 proviral DNA excision using an evolved recombinase. Science.
316(5833): 1912-1915
c. Regenerative medicine
The discovery of the formula -- the so-called Yamanaka’s cocktail -- to generate induced pluripotent stem cells (iPSCs) has opened up myriad possibilities for regenerative medicine, a branch of research that deals with treating diseases through stimulating organs to heal themselves, or to grow organs and tissues in vitro from sources such as stem cells. However, the process of creating iPSCs involves introducing several potential oncogenes into the somatic cells, causing concerns over its safety in clinical use. Fortunately, these transgenes can be removed efficiently using SSR, and this technique has been widely adopted to treat diseases such as Duchenne muscular dystrophy (DMD) and 𝛽-thalassemia.
Examples:
(1) Zhao, C., Farruggio, A.P., Bjornson, C.R.R., Chavez, C.L., Geisinger,
J.M., Neal, T.L., Karow, M., Calos, M.P. (2014) Recombinase-mediated reprogramming and
dystrophin gene addition in mdx mouse induced pluripotent stem cells. PLoS ONE. 9(4):
e96279
(2) Fan, Y., Luo ,Y., Chen, X., Li, Q., Sun, W. (2012) Generation of Human 𝛽-thalassemia
induced pluripotent stem cells from amniotic fluid cells using a single excisable lentiviral
stem cell cassette. Journal of Reproduction and Development. 58(4): 404-409
Why are we creating a modular toolkit for SSR?
SSR is ideal for genetic engineering and synthetic biology
Site-specific recombinases are highly specific. Recognizing relatively long target sites, site-specific recombinases minimize the extent of off-target effects, as longer target sites are less likely to be found in the genome by coincidence. This makes them fit for synthetic biology purposes on top of genetic engineering, as their highly predictable activity allows one to create genetic constructs in a bottom-up way. Considerable effort is also put into protein engineering to minimize off-target effects in certain chassis, and into the discovery of new site-specific recombinases.
Site-specific recombinases are also very simple to use. Unlike other genetic engineering methods such as CRISPR/Cas system, TALEN and ZFN nucleases, site-specific recombinases require only the recombinase protein and two target sites (and occasionally some extra protein factors), and does not require any host processes for genetic engineering [4]. On the contrary, all the aforementioned systems rely on the host DNA repair and homologous recombination machinery to perform knockout and knockin, in which the mechanisms are likely to be error-prone, and are different from species to species. Indeed, site-specific recombinases have been shown to be able to perform genetic engineering in all domains of life, whereas the CRISPR/Cas system cannot be readily adopted in prokaryotes, because prokaryotes generally lack robust DNA repair systems that are found in eukaryotes. This highlights how site-specific recombinase could be superior to alternative genetic engineering methods in terms of organismal flexibility.
SSR performs novel function unachievable in other genetic engineering systems
SSR, unlike other genetic engineering systems, can make dynamic changes to genetic constructs. It is capable of catalyzing not only excision, but also integration, translocation, and inversion, which are highly inefficient and difficult, if not impossible, to perform using other genetic engineering systems. These special properties have allowed researchers to build novel devices with applications ranging from studying brain function through tagging individual neurons , to speeding up bone healing by gene therapy.
More importantly, the existence of many orthogonal site-specific recombinases means that multiple genetic engineering can occur in parallel without affecting each other. They have been employed in efficient genetic assembly , as well as the creation of a six-input biological logic gate . In our project, we also demonstrated the utility of multiple orthogonal systems through gene randomiser, logic gates, and pulse generator .
SSR complements other genetic engineering technology
Although SSR itself is already a powerful genetic tool, its functionality can be greatly expanded when used in conjunction with tools such as CRISPR/Cas9. One of the major inconveniences of using SSR is the need to introduce a target site into the genome of the organism; this could be easily solved by CRISPR/Cas9, which induces homology-directed repair (HDR) to integrate transgene into the host genome. On the other hand, CRISPR/Cas9 may not be the best tool when fine spatiotemporal control is required; SSR would be more suitable in this case.
It is therefore not surprising that researchers are already using SSR and CRISPR/Cas9 together in experiments. They are already used in applications such as engineering stem cells [5], speeding up vaccine development [6], studying cancer mechanism [7] and parasite infection [8], and more.
SSR technology is not fully exploited
While researchers have benefitted enormously from the development of a few recombinases (Cre and FLP being the prime examples), we realise that the potential of SSR in genetic engineering is yet to be fully exploited because of the limited choices of well-characterized recombinases. The activity of recombinases varies between in vitro and in vivo conditions, between organisms, and even between tissue types. For example, Cre recombinase is shown to be toxic in mouse lungs and hearts. The limited choices of recombinases and their incomprehensive testing put restrictions to experimental design, ultimately hindering scientific discovery
By developing a site-specific, modular recombination (SMORE) toolkit with each part thoroughly tested, we aim to contribute multiple usable recombination systems to the scientific community, allowing the creation of complex gene circuits for various applications. We envision that SMORE will facilitate the use of multiple orthogonal SSR in vivo and in parallel, with applications such as multiple inducible gene knockout, as well as in the field of metabolic engineering, which we plan to demonstrate.
What does SMORE contain?

As a toolkit, SMORE contains individual recombinases expression constructs and their respective target sites in biobrick format. Following the principles of synthetic biology, we endeavor to characterise all of our parts as thoroughly as possible, through in vivo or in vitro testing. This is why we have included constructs in the SMORE toolkit for testing their activity, which lays out the framework for the characterization of recombinase activity. To learn more about the characterisation of individual parts, please check our results and parts pages.
Additionally, we are aware of the emerging role of computational modeling in synthetic biology, and we have used computer simulation extensively to predict the behaviour of recombination. Click here to learn more about our modeling.
Lastly, we believe that synthetic biology should be accessible to people from all disciplines. This is why we have chosen to investigate on ‘accessibility’ as our human practice. Click here to see more about our findings!
References
[1] Leach, D. Genetic recombination. Oxford: Blackwell Science 1996
[2] Kamisugi, Y., Schlink, K., Rensing, S.A., Schween, G., von Stackelberg, M., Cuming, A.C., Reski, R., Cove, D.J. (2006) The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Research. 34(21): 6205-6214
[3] Terada, R., Johzuka-Hisatomi, Y., Saitoh, M., Asao, H., Iida, S. (2007) Gene Targeting by Homologous Recombination as a Biotechnological Tool for Rice Functional Genomics. Plant Physiology. 144(2): 846-856
[4] Grindley, N.D.F., Whiteson, K.L., Rice, P.A. (2006) Mechanisms of site-specific recombination. Annual Review of Biochemistry. 75: 567-605
[5] Chen, Y., Cao, J., Xiong, M., Peterson, A.J., Dong, Y., Tao, Y., Huang, C.T-L., Du, Z., Zhang, S-C. (2015) Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell. 17(2): 233-244
[6] Liang, X., Sun, L., Yu, T., Pan, Y., Wang, D., Hu, X., Fu, Z., He, Q., Cao, G. (2016) A CRISPR/Cas9 and Cre/Lox system-based express vaccine development strategy against re-emerging Pseudorabies virus. Scientific Reports. 19176.
[7] Vanoli, F., Jasin, M. (2017) Generation of chromosomal translocations that lead to conditional fusion protein expression using CRISPR-Cas9 and homology-directed repair. Methods. 121-122: 138-145
[8] Knuepfer, E., Napiorkowska, M., van Ooij, C., Holder, A.A. (2017) Generating conditional gene knockouts in Plasmodium– a toolkit to produce stable DiCre recombinase-expressing parasite lines using CRISPR/Cas9. Scientific Reports. 7: 3881
















