| Line 3: | Line 3: | ||
[[File:T--Aix-Marseille--EPSd.png|450px|right|thumb|EPS Depolymerase process to clean the xylem vessel from EPS.]] | [[File:T--Aix-Marseille--EPSd.png|450px|right|thumb|EPS Depolymerase process to clean the xylem vessel from EPS.]] | ||
| − | To fight | + | To fight [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']] we searched for natural solutions. |
| + | Some phages have a devious way to attack bacteria, digesting the biofilm so that they can their target more easily <ref>Vandenbergh, P. A., Wright, A. M. & Vidaver, A. K. Partial Purification and Characterization of a Polysaccharide Depolymerase Associated with Phage-Infected Erwinia amylovora. Appl. Environ. Microbiol. 49, 994–996 (1985)</ref>. | ||
| + | We decided to try to use this approach to attack [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']]. | ||
| + | We found an enzyme called EPS-depolymerase <ref>Kim, W. S. & Geider, K. Characterization of a Viral EPS-Depolymerase, a Potential Tool for Control of Fire Blight. Phytopathology 90, 1263–1268 (2000).</ref> that can hydrolyze the Exopolysaccharide (EPS) forming the biofilm. | ||
| − | As the symptoms observed in plants are the result | + | As the symptoms observed in plants are the result occlusion of xylem vessels by bacterial biofilm and the accumulation of EPS, |
| − | + | we think this may be a way to unblock the vessels. | |
| − | + | The goal of part of the design isn’t to treat [[Team:Aix-Marseille/Xylella_fastidiosa|''X. fastidiosa'']] or to prevent the infection, but to reduce symptoms. | |
==EPS Depolymerase design== | ==EPS Depolymerase design== | ||
| Line 13: | Line 16: | ||
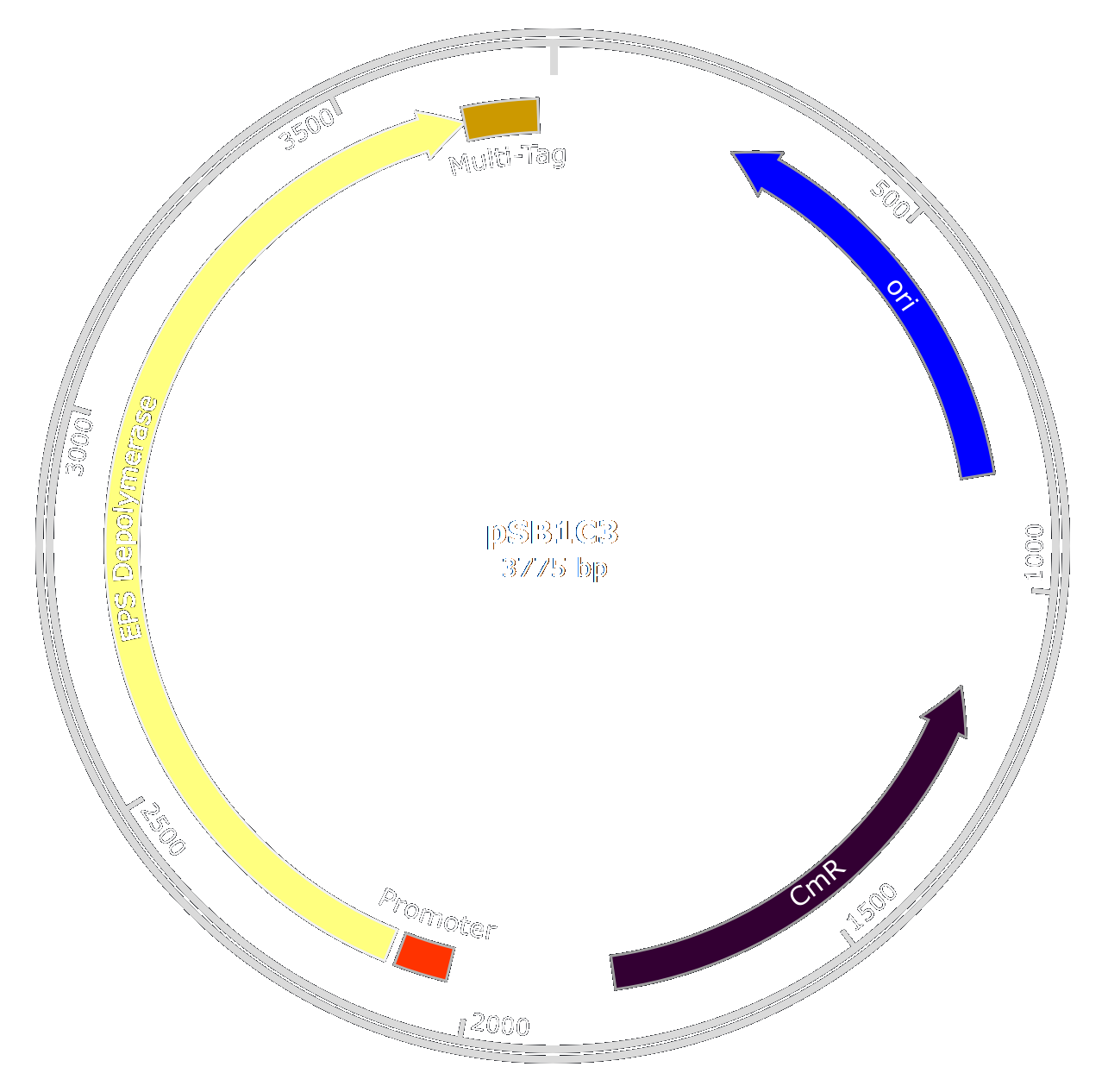
[[File:T--Aix-Marseille--EPSplasmid-2.png|450px|right|thumb]] | [[File:T--Aix-Marseille--EPSplasmid-2.png|450px|right|thumb]] | ||
| − | As we wanted to limit the number of GMOs in our product | + | As we wanted to limit the number of GMOs in our product decided to purify the enzyme after production in ''E. coli'' using a quick and efficient purification system. |
| − | The original enzyme used by the phage is | + | The original enzyme used by the phage is very large and the gene is not optimal for a production in ''E. coli''. |
| + | Thus, to produce large quantities of the enzyme, we decided to optimize the enzyme by taking only the catalytic domain of the original protein and optimizing the gene sequence for production in ''E. coli''. | ||
| + | This lead to the creation of the EPS-Depolymerase part : [http://parts.igem.org/Part:BBa_K2255006 BBa_K2255006]. | ||
| − | As we wanted to purify the EPS Depolymerase | + | As we wanted to purify the EPS Depolymerase to use it in our treatment, we also engineered a multi-tag. |
| + | This tag is composed of an oligo-histidine-tag, that binds to Nickel or Cobalt ions, and a Strep-tag that binds to streptavidin separated by a TEV cleavage site. | ||
| + | Thus we created the biobrick [http://parts.igem.org/Part:BBa_K2255003 BBa_K2255003]. | ||
| − | + | The two biobricks will be fused using the [http://parts.igem.org/Assembly_standard_25 Rfc25] standard to produce an easilly purifiable enzyme. | |
| + | To get large amounts, we decided to add a strong and constitutive promoter and RBS in ''E.coli'' ([http://parts.igem.org/Part:BBa_K608002 BBa_K608002]). | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 11:55, 1 November 2017
EPS Depolymerase
Contents
To fight Xylella fastidiosa we searched for natural solutions. Some phages have a devious way to attack bacteria, digesting the biofilm so that they can their target more easily [1]. We decided to try to use this approach to attack Xylella fastidiosa. We found an enzyme called EPS-depolymerase [2] that can hydrolyze the Exopolysaccharide (EPS) forming the biofilm.
As the symptoms observed in plants are the result occlusion of xylem vessels by bacterial biofilm and the accumulation of EPS, we think this may be a way to unblock the vessels. The goal of part of the design isn’t to treat X. fastidiosa or to prevent the infection, but to reduce symptoms.
EPS Depolymerase design
As we wanted to limit the number of GMOs in our product decided to purify the enzyme after production in E. coli using a quick and efficient purification system.
The original enzyme used by the phage is very large and the gene is not optimal for a production in E. coli. Thus, to produce large quantities of the enzyme, we decided to optimize the enzyme by taking only the catalytic domain of the original protein and optimizing the gene sequence for production in E. coli. This lead to the creation of the EPS-Depolymerase part : [http://parts.igem.org/Part:BBa_K2255006 BBa_K2255006].
As we wanted to purify the EPS Depolymerase to use it in our treatment, we also engineered a multi-tag. This tag is composed of an oligo-histidine-tag, that binds to Nickel or Cobalt ions, and a Strep-tag that binds to streptavidin separated by a TEV cleavage site. Thus we created the biobrick [http://parts.igem.org/Part:BBa_K2255003 BBa_K2255003].
The two biobricks will be fused using the [http://parts.igem.org/Assembly_standard_25 Rfc25] standard to produce an easilly purifiable enzyme. To get large amounts, we decided to add a strong and constitutive promoter and RBS in E.coli ([http://parts.igem.org/Part:BBa_K608002 BBa_K608002]).
References
- ↑ Vandenbergh, P. A., Wright, A. M. & Vidaver, A. K. Partial Purification and Characterization of a Polysaccharide Depolymerase Associated with Phage-Infected Erwinia amylovora. Appl. Environ. Microbiol. 49, 994–996 (1985)
- ↑ Kim, W. S. & Geider, K. Characterization of a Viral EPS-Depolymerase, a Potential Tool for Control of Fire Blight. Phytopathology 90, 1263–1268 (2000).