(→How close can the numbers be when fluorescence is measured all around the world ?) |
(→Materials and Methods:) |
||
| Line 12: | Line 12: | ||
==Materials and Methods:== | ==Materials and Methods:== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[File:T--Aix-Marseille--fluo interlab.jpeg|right]] | [[File:T--Aix-Marseille--fluo interlab.jpeg|right]] | ||
| − | + | InterLab Measurement Study has two components: calibration and cell measurement. The calibration component aims to identify the optimal setting for the plate reader to read the OD600 and fluorescence. We used plasmids containing different promoter from the 2017 distribution kit and transfected them into ''E.coli''. | |
| − | + | ||
| − | + | ||
| − | + | Before measuring fluorescence expression, we used LUDOX and fluorescein to calibrate the TECAN. Also, we prepared a dilution series of fluorescein in 4 duplicates and measure the fluorescence in a 96 well plate in a plate reader. By measuring these in all standard modes, we generated a standard curve of fluorescence for fluorescein concentration. We used this to correct cell based readings to an equivalent fluorescein concentration and convert this into a GFP concentration. | |
| − | + | ||
| + | More details on the [https://static.igem.org/mediawiki/2017/8/85/InterLab_2017_Plate_Reader_Protocol.pdf iGEM Protocol]. | ||
[[File:T--Aix-Marseille--fluoresceincurve.jpeg|500px|center]] | [[File:T--Aix-Marseille--fluoresceincurve.jpeg|500px|center]] | ||
Revision as of 21:02, 1 November 2017
InterLab
How close can the numbers be when fluorescence is measured all around the world ?
In order to support the interlab study, our team participated in the Fourth International InterLab Measurement Study. We were really happy to contribute to this study based on reliable and repeatable measurements. The challenge this year consisted in establishing a GFP measurement protocol that can be used produce comparable GFP measurements on plate readers. This study aims to mesure fluorescence value from the E. coli K-12 DH5α cells transformed with different plasmids, carrying GFP genes un der the control of various promotors. Furthermore, teams also tested some RBS devices that are intended to make gene expression more precise and reliable.
Materials and Methods:
InterLab Measurement Study has two components: calibration and cell measurement. The calibration component aims to identify the optimal setting for the plate reader to read the OD600 and fluorescence. We used plasmids containing different promoter from the 2017 distribution kit and transfected them into E.coli.
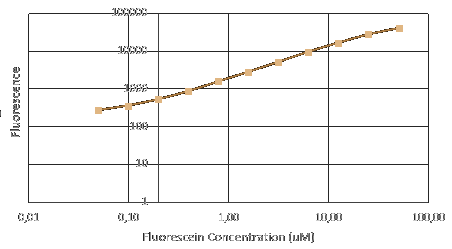
Before measuring fluorescence expression, we used LUDOX and fluorescein to calibrate the TECAN. Also, we prepared a dilution series of fluorescein in 4 duplicates and measure the fluorescence in a 96 well plate in a plate reader. By measuring these in all standard modes, we generated a standard curve of fluorescence for fluorescein concentration. We used this to correct cell based readings to an equivalent fluorescein concentration and convert this into a GFP concentration.
More details on the iGEM Protocol.
Results
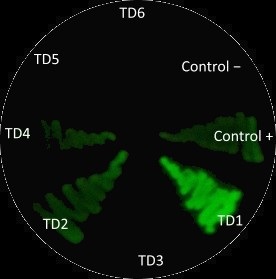
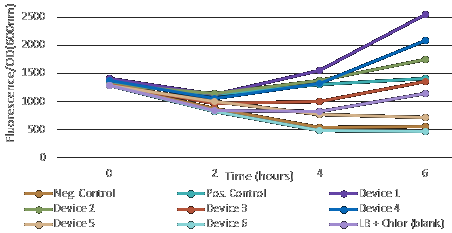
After measuring fluorescence readings and OD, we calculed ratio of Fluorescence/OD in order to determine promotor strengh. Device 1 and device 4 containing promoter J23101 gave high fluorescence measurements.
Device 2 (J23106) and device 3 (J23117) were observed to have a medium increase of fluorescence.
However, device 5 (J23106) and device 6 (J23117) showed low fluorescence.
This results suggest that the promoter J23101 had the strongest affinity for the RNA polymerase when compared with other promoters J23117 and J23106.
In addition, we interessing in RBS devices (BCDs) that are intended to make gene expression more precise and reliable. Devices 1, 2 and 3 contain the rbs B0034, while devices 4, 5 and 6 contain the rbs BCD2. That can explain experimental results, regarding to theoretical strengh for promotors. For example, we observe a higher GFP signal for device 3 than device 5, despite of an stronger promotor for device 5.
Promotor strength(iGEM data):
- J23101(TD1,TD4):1791
- J23106(TD2,TD5):1185
- J23117(TD3,TD6):162
Overall, we can supposed that device 1 and 4 have the strongest promotor. The point on RBS is that B0034 owns an higher specificity, explaining why device 1 is higher thant device 4.