(→Competent bacteria:) |
(→Competent bacteria:) |
||
| Line 61: | Line 61: | ||
Then we Made two solutions necessary for making the competent cells. | Then we Made two solutions necessary for making the competent cells. | ||
$$ \textit{Buffer 1} - \text{ 80 mL: } | $$ \textit{Buffer 1} - \text{ 80 mL: } | ||
| − | \text{ 2.4 } \text{ mL } \ce{KAc} \text{ [1 M] }, \ | + | \text{ 2.4 } \text{ mL } \ce{KAc} \text{ [1 M] }, \pu{8 mL } \ce{ MnCl2 } \text{ [0.5 M] } , \text{8 mL} \ce{ KCl } \text{ [1 M] } , \text{ 8 mL } \ce{ CaCl2 } \text{ [0.1 M] }, \text{ 15 mL } \text{ Gly } \text{ [80%] } , \text{ → } \text{ 38.6 mL } \ce{ H2O } $$ |
$$ \textit{Buffer 2} - \text{8 mL:} | $$ \textit{Buffer 2} - \text{8 mL:} | ||
\text{ 400 } \text{ μL } \text{ MOPS } \text{ [0.2M] }, \text{ 6 mL } \ce{ CaCl2 } \text{ [0.1 M] }, \text{ 1.5 } \text{ mL } | \text{ 400 } \text{ μL } \text{ MOPS } \text{ [0.2M] }, \text{ 6 mL } \ce{ CaCl2 } \text{ [0.1 M] }, \text{ 1.5 } \text{ mL } | ||
Revision as of 10:55, 16 June 2017
{{{title}}}
CALENDRIER DESACTIVE EN ATTENDANT DE FINIR lA RETRANSCRIPTION
Day 1 : 06/06/2017
Cleaning the laboratory, then recovery and installation of the equipment. Starting culture of strains TG1 and DH5α. We launched precultures from cryotubes ( sampling with 1 rod and then deposition in an erlenmeyer containing LB media ). Incubation at 37 ° C.
Day 2 : 07/06/2017
Strains cultivation:
Recovery of strains cultivated the day before: DH5α and TG1 To check the concentrations, the OD is measured. For doing so, the culture must be diluted because the optimum measurement of the apparatus is between 0.1 and 0.8. The cultures in the tank are diluted to 1/10 with distilled water. $$\text{ Our results } : \text{ TG1: 5.16 } / \text{ DH5α: 5.21 }$$ To calculate the volume to be taken for our 100 mL we do: $$\frac{\text{OD wanted}}{\text{OD obtained before dilution}} \times \text{Volume} = \frac{0.1}{5.16} \times 100 \simeq 2 \text{ mL}$$ E. coli being aerobic, the solution is stored in LB in a Erlenmeyer flask of 5 × volume, so 500 mL and incubate for 1 h.
Glycerolization of strains:
Putting the strains in glycerol protect them from the cold, necessary when one will freeze at -80 ° C so that the cells do not collapse. 40% Glycerol in 1.8mL cryotubes → 0.9 of glycerol and 0.9 of strains. We will freeze the cells in exponential phases in order to make our cells competent, ie able to incorporate DNA.
Competent bacteria:
To make competent bacteria, it is necessary to take them in exponential growth phase. A quantity of bacterium is thus taken which is placed in LB medium and then incubated for 1 hour at 37 degree Celsius. An OD value of 0.1 is desired in 100 mL. For TG1-> 1.94 mL For DH5α-> 1.92 mL
Mesuring OD after 1h40:
TG1 - 0.7
DH5α - 0.5
The cultures were thus recovered and were divided into falcons (50 ml each) and placed in ice. (For each of the two strains).
Preparation of the buffers: Solution TBF1 and TBF2
Preparation of buffer solutions:
KAc: 50 mL → 4.9 g
MnCl2: 200 mL → 19.79 g
KCl: 200 mL → 14.91 g
MOPS: 50 mL → 2.09 g
Each solution was autoclaved.
Then we Made two solutions necessary for making the competent cells.
$$ \textit{Buffer 1} - \text{ 80 mL: }
\text{ 2.4 } \text{ mL } \ce{KAc} \text{ [1 M] }, \pu{8 mL } \ce{ MnCl2 } \text{ [0.5 M] } , \text{8 mL} \ce{ KCl } \text{ [1 M] } , \text{ 8 mL } \ce{ CaCl2 } \text{ [0.1 M] }, \text{ 15 mL } \text{ Gly } \text{ [80%] } , \text{ → } \text{ 38.6 mL } \ce{ H2O } $$
$$ \textit{Buffer 2} - \text{8 mL:}
\text{ 400 } \text{ μL } \text{ MOPS } \text{ [0.2M] }, \text{ 6 mL } \ce{ CaCl2 } \text{ [0.1 M] }, \text{ 1.5 } \text{ mL }
\text{ Gly } \text{ [80%] }, \text{ 80 μL } \ce{ KCl } \text{ [1 M] }, \text{ 500 μL } \ce{ H2O}$$
To make competent cells, we ALWAYS work at low temperature (4 ° C) and in sterile medium. The buffers thus prepared were stored in ice.
Centrifugation of the cell culture 10 min at 3500 rpm at 4 ° C. The pellet must then be resuspended (gently) in 80 ml of Tbf1 (20 ml as 50 ml of culture in each falcon).
3500 rpm centrifugation for 5 minutes.
Then resuspension of the pellet in 2 mL of Tbf2.
Let incubate for 15 minutes in ice and then aliquot 200 μL of bacteria solution in a cryotube and store at -80 ° C. These aliquots are at I8 for competent DH5 alpha and I9 for competent TG1.
The remainder is stored at -20 ° C in labeled falcons.
After the first centrifugation was carried out in a cold (non-sterile) chamber, we re-started precultures from those in the morning: 100 μL of culture were placed in 10 ml of LB medium and then incubated at 37 ° C. overnight.
The buffer Tbf1 and Tbf2 have also been prepared to be ready so we can restart the manipets in case of failure.
Day 3: 08/06/17
Preparation for transformation : In four steps
For the petri dish LB agar, take a bottle of LB agar ( 400mL for 20 boxes ) and put it in the microwave to liquefy. Heat with microwave 300 watt ( no more ) for 19 minutes with unscrewed plug.
Organizational axis
Subjects organisation
| Killer red | Crispr Cas9 | P3 | Génome M13 | Measurement | DEPS | QS |
| Robin | Flora/Soraya | Camille | Camille/Thibault | Lisa | Hussein | Jeremy Flora Ilann |
Small theoretical session of Sandra on the manipulations:
1. Transformation
Transformation is a technic used to make the DNA "enter" into the competent cell by destabilizing the membrane:
- Contact, the cells and plasmids (which are recovered in the iGEM kit)
- Heat shock, more permeable membrane that will make the DNA enter: 45 seconds at 42 ° C
- Expression, cells are allowed to express the inserted genes (including antibiotic resistance which serve as a test). There are two types of antibiotics: bacteriostatic (ampicillin, which freezes growth), bactericidal (which kills)
- Spread on antibiotic (petri dish), that will eliminate the bacteria which have not assimilated the gene
2. Verification of contamination
Negative control: 3 antibiotics, if the bacteria resist: contaminated
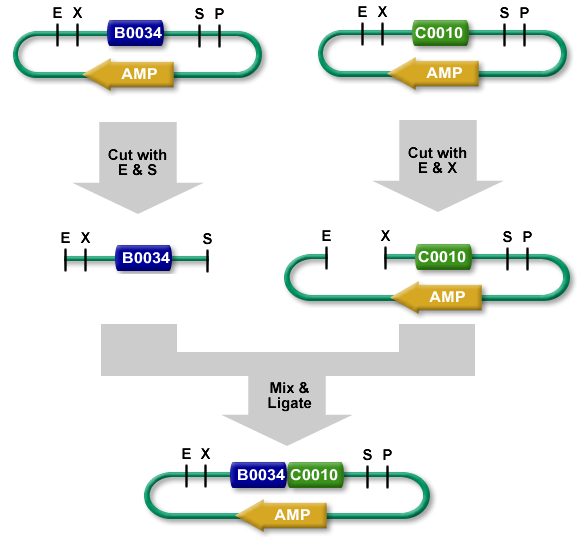
3. Cloning
We want to integrate an insert, for this, we need the restriction enzymes
4. Biobrick - iGEM
S and X are compatible
Day 4: 09/06/17
BROUILLON
B. Re-culture
In order to have uncontaminated crops (cultures - -) in case the one made yesterday were. C. Biobrick Putting GFP into solution from the iGEM Kit (Item 5: 16K): 10 μL of H2O miliQ (pure without DNases or RNAses) are deposited and incubated for 10 minutes. The 10 μL of DNA is recovered taking care to take the edge of the cuvée.
Opening of the stopper (in the loom) 19min to 300W MAXIMUM and left at 50 ° C in an incubator.
Recovery of antibiotics and dosages. Here are the different products to respect Ampicillin -> 4 μL / mL Ampi [25mg / mL] -> here 320μL Kanamycin -> 5 μL / mL Kana [10mg / mL] -> here 400 μL Tetracycline -> 1 μL / mL Tetra [15mg / mL] -> here at 80μL Chloramphenicol -> 1.6 μL / mL Chloran [30mg / mL] here 128μL
(Re) preparation of competent bacterial cells: Error of the previous day.
Recovery of precision and OD measurements from diluted sample to tenth: OD TG1 = 5.24 OD DH5α = 4.27
A quantity of bacterium is then taken which has been placed in an LB medium and then incubated for approximately 1 hour at 37 ° C. 1.9 mL of TG1 in 100 mL of LB 2.3 mL of DH5α in 100 mL of LB Then measurement of OD TG1 = 0.474 / DH5a = 0.895 (a little high compared to the desired value).
To make competent cells, it is imperative to work at low temperature (4 ° C) and in sterile medium. The buffers thus prepared were kept and the eppendorf tubes must be cold (cold room and ice cube tray). First, 10 min centrifugation at 3500 rpm at 4 ° C. of the two bacterial strains from the culture (OD 0.4-0.6). The pellet should then be gently resuspended in 80 mL of Tbf1 (20 mL in our case because we do not have 200 mL of culture). 3500 rpm centrifugation with cold pendant 5 mn and resuspension of the new pellet in 2 mL of Tbf2. Incubation of 15mn in ice. Manufacture of 220μL aliquots in annotated eppendorfs STERILE and bath tubes in liquid nitrogen to freeze them quickly. Storage at -80 ° C
Jour n°7. 14/06/17
Digestion vector:
We prepared 6 eppendorf tubes, we add in first the buffer ( with the same cone ) and H2O ( with a second cone ). Afterward we add the DNA material for a total mass of 200 ng in each tube. When the initial preparation was done, a short spin was necessary. We extracted the enzyme carefully with a small up action on the edge of the enzymes tubes. Then carefully inject the enzymes in the eppendorf tubes then mix with an up down action. Then we incubate for half an hour at 37° degrees celsius. Afterward, a heat shock is applied at 80°C for 20 minutes to inactivate all enzyme activity.
Tableau / code
Digestion plasmid:
We had 4 tubes of plasmid with each a specific antibiotic resistant gene insert. With a total of volume of 45µL in each. We added 10µL of NE buffer, 41µL of H2O at the end we added our enzymes. ( 2µL for ECORI HF and 2µL for PST1 ). Then we incubate for half an hour at 37° degrees celsius. Afterward, a heat shock is applied at 80°C for 20 minutes to inactivate all enzyme activity.
Ligation:
We prepared 3 tubes for ligation for which we added 2 µL of T4 DNA ligase buffer 10x. Then 11µL of ddH2O, afterward the DNA material ( 2µL upstream 2µL downstream 2µL destination plasmid ). Finally we add T4 DNA ligase and let it incubate at room temperature for 10mn. When the ligation was done, we transformed 5µL of ligation product into 150µL of bacterial cell. ( Previously mentioned ).
Electrophoresis:
As mentioned previously, for each ligation product we added in an eppendorf tube 5µL of DNA 10µL of H2O, 3µL of loading purple dye. In the gel, the order was as is EPRI cut up, EPRC cut up, EPRC cut down, DNA ladder, RBS cut down, GFP cut down, M13 cut up. We let it migrate for 30 minutes at 135 volt? Which was the limit of the migration, we should have put it only for 25. All fragment under 200 Pb, aren’t observed because it had surpassed the limit of migration. So for EPRI, EPRC cut up and down, we should only see one bar for the linear plasmid and not for the digested factors.