(→Day 16) |
SuperFlower (Talk | contribs) |
||
| Line 295: | Line 295: | ||
We prepared 3 tubes for ligation for which we added 2 µL of T4 DNA ligase buffer 10x. Then 11µL of ddH2O, afterward the DNA material ( 2µL upstream 2µL downstream 2µL destination plasmid ). Finally we add T4 DNA ligase and let it incubate at room temperature for 10mn. | We prepared 3 tubes for ligation for which we added 2 µL of T4 DNA ligase buffer 10x. Then 11µL of ddH2O, afterward the DNA material ( 2µL upstream 2µL downstream 2µL destination plasmid ). Finally we add T4 DNA ligase and let it incubate at room temperature for 10mn. | ||
When the ligation was done, we transformed 5µL of ligation product into 150µL of bacterial cell. ( Previously mentioned ). | When the ligation was done, we transformed 5µL of ligation product into 150µL of bacterial cell. ( Previously mentioned ). | ||
| + | |||
| + | |||
| + | ----------------------------------------------------------------------------------------------------------------------- | ||
| + | |||
Electrophoresis: | Electrophoresis: | ||
| − | As mentioned previously, for each ligation product we added in an eppendorf tube 5µL of DNA 10µL of H2O, 3µL of loading purple dye. In the gel, the order was | + | As mentioned previously, for each ligation product we added in an eppendorf tube 5µL of DNA 10µL of H2O, 3µL of loading purple dye. In the gel, the order was EPRI cut up, EPRC cut up, EPRC cut down, DNA ladder, RBS cut down, GFP cut down, M13 cut up. |
| − | We let it migrate for 30 minutes at 135 volt | + | We let it migrate for than 30 minutes, at 135 volt which was the limit of the migration, we should have put it for only 25. |
| − | All fragment under 200 Pb | + | All fragment under 200 Pb weren't displayed because it had surpassed the migration cap. |
| − | + | For EPRI, EPRC cut up and down, we should have seen one bar for the linear plasmid and not for the digested factors. | |
PHOTO ELECTRO | PHOTO ELECTRO | ||
| − | We also re-did a transformation for RBS which failed previously. This time we used new competent cells and we had a positive result because there | + | |
| + | We also re-did a transformation for RBS which failed previously. This time we used new competent cells and we had a positive result because there were multiple colonies on the gel. | ||
==DAY 8: 15/06/17== | ==DAY 8: 15/06/17== | ||
| Line 423: | Line 428: | ||
Afterward, 20µL of normal H2O. A centri for 1mn at 30 to 50g then another at 11kg. We repeated two times and kept the flow through. We measured the DNA concentration using nanodrop the values mentioned as is. | Afterward, 20µL of normal H2O. A centri for 1mn at 30 to 50g then another at 11kg. We repeated two times and kept the flow through. We measured the DNA concentration using nanodrop the values mentioned as is. | ||
| − | + | Cas9 -> 4,48ng/µL | |
| + | dcas9 -> 4,24ng/µL | ||
| + | 8B -> 9,54ng/µL | ||
| + | 14B -> 11,88ng/µL | ||
| + | |||
We also did a PCR for Cas9 and dCas9. | We also did a PCR for Cas9 and dCas9. | ||
| Line 437: | Line 446: | ||
Transplantation of 4 plates of phage from 20/01 in a Kanamicin Petri Dish. | Transplantation of 4 plates of phage from 20/01 in a Kanamicin Petri Dish. | ||
| − | + | ||
| + | The competence test reveled that our super competent bacteria are approximately three times more competent than Sandra's one, and approximately 1,5 times more competent than | ||
Le test de compétence a révélé que nos bactéries super compétente sont en moyenne 3 fois plus compétentes que les compétentes de Sandra, et environ 1.5 fois plus compétentes que les super compétentes de l’équipe voisine. | Le test de compétence a révélé que nos bactéries super compétente sont en moyenne 3 fois plus compétentes que les compétentes de Sandra, et environ 1.5 fois plus compétentes que les super compétentes de l’équipe voisine. | ||
| Line 474: | Line 484: | ||
On the petri dish with GM1 E.coli infected with M13 phage, coming from the 20/06, we seen multiple infection site. Thus we transferred 4 of them to a kanamycin petri dish. | On the petri dish with GM1 E.coli infected with M13 phage, coming from the 20/06, we seen multiple infection site. Thus we transferred 4 of them to a kanamycin petri dish. | ||
| + | |||
==Day 14 : 22/06== | ==Day 14 : 22/06== | ||
Revision as of 14:52, 6 July 2017
{{{title}}}
CALENDRIER DESACTIVE EN ATTENDANT DE FINIR lA RETRANSCRIPTION
Contents
Day 1 : 06/06/2017
Cleaning the laboratory, then recovery and installation of the equipment. Starting culture of strains TG1 and DH5α. We launched precultures from cryotubes ( sampling with 1 rod and then deposition in an erlenmeyer containing LB media ). Incubation at 37 ° C.
Day 2 : 07/06/2017
Strains cultivation:
Recovery of strains cultivated the day before: DH5α and TG1 To check the concentrations, the OD is measured. For doing so, the culture must be diluted because the optimum measurement of the apparatus is between 0.1 and 0.8. The cultures in the tank are diluted to 1/10 with distilled water.
Our results : TG1: 5.16 / DH5α: 5.21
To calculate the volume to be taken for our 100 mL we do: $$\frac{\text{OD wanted}}{\text{OD obtained before dilution}} \times \text{Volume} = \frac{0.1}{5.16} \times 100 \simeq 2 \text{ mL}$$ E. coli being aerobic, the solution is stored in LB in a Erlenmeyer flask of 5 × volume, so 500 mL and incubate for 1 h.
Glycerolization of strains:
Putting the strains in glycerol protect them from the cold, necessary when one will freeze at -80 ° C so that the cells do not collapse. 40% Glycerol in 1.8mL cryotubes → 0.9 of glycerol and 0.9 of strains. We will freeze the cells in exponential phases in order to make our cells competent, ie able to incorporate DNA.
Competent bacteria:
To make competent bacteria, it is necessary to take them in exponential growth phase. A quantity of bacterium is thus taken which is placed in LB medium and then incubated for 1 hour at 37 degree Celsius. An OD value of 0.1 is desired in 100 mL. For TG1-> 1.94 mL For DH5α-> 1.92 mL
Mesuring OD after 1h40:
TG1 - 0.7
DH5α - 0.5
The cultures were thus recovered and were divided into falcons (50 ml each) and placed in ice. (For each of the two strains).
Preparation of the buffers: Solution TBF1 and TBF2
Preparation of buffer solutions:
KAc: 50 mL → 4.9 g
MnCl2: 200 mL → 19.79 g
KCl: 200 mL → 14.91 g
MOPS: 50 mL → 2.09 g
Each solution was autoclaved.
Then we made two solutions necessary for making the competent cells.
To make competent cells, we ALWAYS work at low temperature (4 ° C) and in sterile medium. The buffers thus prepared were stored in ice. Centrifugation of the cell culture 10 min at 3500 rpm at 4 ° C. The pellet must then be resuspended (gently) in 80 ml of Tbf1 (20 ml as 50 ml of culture in each falcon). 3500 rpm centrifugation for 5 minutes.
Then resuspension of the pellet in 2 mL of Tbf2.
Let incubate for 15 minutes in ice and then aliquot 200 μL of bacteria solution in a cryotube and store at -80 ° C. These aliquots are at I8 for competent DH5 alpha and I9 for competent TG1.
The remainder is stored at -20 ° C in labeled falcons.
After the first centrifugation was carried out in a cold (non-sterile) chamber, we re-started precultures from those in the morning: 100 μL of culture were placed in 10 ml of LB medium and then incubated at 37 ° C. overnight.
The buffer Tbf1 and Tbf2 have also been prepared to be ready so we can restart the manipets in case of failure.
Day 3: 08/06/17
Preparation for transformation : In four steps
For the petri dish LB agar, take a bottle of LB agar ( 400mL for 20 boxes ) and put it in the microwave to liquefy. Heat with microwave 300 watt ( no more ) for 19 minutes with unscrewed plug.
Organizational axis
Subjects organisation
| Killer red | Crispr Cas9 | P3 | Génome M13 | Measurement | DEPS | QS |
| Robin | Flora/Soraya | Camille | Camille/Thibault | Lisa | Hussein | Jeremy Flora Ilann |
Small theoretical session of Sandra on the manipulations:
1. Transformation
Transformation is a technic used to make the DNA "enter" into the competent cell by destabilizing the membrane:
- Contact, the cells and plasmids (which are recovered in the iGEM kit)
- Heat shock, more permeable membrane that will make the DNA enter: 45 seconds at 42 ° C
- Expression, cells are allowed to express the inserted genes (including antibiotic resistance which serve as a test). There are two types of antibiotics: bacteriostatic (ampicillin, which freezes growth), bactericidal (which kills)
- Spread on antibiotic (petri dish), that will eliminate the bacteria which have not assimilated the gene
2. Verification of contamination
Negative control: 3 antibiotics, if the bacteria resist: contaminated
3. Cloning
We want to integrate an insert, for this, we need the restriction enzymes
4. Biobrick - iGEM
S and X are compatible
Day 4: 09/06/17
BROUILLON
B. Re-culture
In order to have uncontaminated crops (cultures - -) in case the one made yesterday were. C. Biobrick Putting GFP into solution from the iGEM Kit (Item 5: 16K): 10 μL of H2O miliQ (pure without DNases or RNAses) are deposited and incubated for 10 minutes. The 10 μL of DNA is recovered taking care to take the edge of the cuvée.
Opening of the stopper (in the loom) 19min to 300W MAXIMUM and left at 50 ° C in an incubator.
Recovery of antibiotics and dosages. Here are the different products to respect Ampicillin -> 4 μL / mL Ampi [25mg / mL] -> here 320μL Kanamycin -> 5 μL / mL Kana [10mg / mL] -> here 400 μL Tetracycline -> 1 μL / mL Tetra [15mg / mL] -> here at 80μL Chloramphenicol -> 1.6 μL / mL Chloran [30mg / mL] here 128μL
(Re) preparation of competent bacterial cells: Error of the previous day.
Recovery of precision and OD measurements from diluted sample to tenth: OD TG1 = 5.24 OD DH5α = 4.27
A quantity of bacterium is then taken which has been placed in an LB medium and then incubated for approximately 1 hour at 37 ° C. 1.9 mL of TG1 in 100 mL of LB 2.3 mL of DH5α in 100 mL of LB Then measurement of OD TG1 = 0.474 / DH5a = 0.895 (a little high compared to the desired value).
To make competent cells, it is imperative to work at low temperature (4 ° C) and in sterile medium. The buffers thus prepared were kept and the eppendorf tubes must be cold (cold room and ice cube tray). First, 10 min centrifugation at 3500 rpm at 4 ° C. of the two bacterial strains from the culture (OD 0.4-0.6). The pellet should then be gently resuspended in 80 mL of Tbf1 (20 mL in our case because we do not have 200 mL of culture). 3500 rpm centrifugation with cold pendant 5 mn and resuspension of the new pellet in 2 mL of Tbf2. Incubation of 15mn in ice. Manufacture of 220μL aliquots in annotated eppendorfs STERILE and bath tubes in liquid nitrogen to freeze them quickly. Storage at -80 ° C
DAY 6: 13/06/17
BROUILLON
Mini préparation.
Avant de commencer Sur centrifugeuse 20μL de Lyse bleu et 200μL de Rnase dans les tubes initiaux. Sur l'ajoute 20μL de Lyse blue au buffer P1. (Quantité initiale du tampon P1 = 20 ml). Puis sur rajoute 200μL de Rnase 1 au "tampon P1 + Lyse bleu". La solution est conservée dans la glace. L'éthanol (96-100%) "ajouter au tampon PE avant utilisation" était déjà ajouté. Nous avons donc utilisé le buffer PE.
Debut de la mini préparation. Sur rempli nos tubes ependorfs avec les bactéries conservées dans nos entrées. Dans l'ordre -Tube 1: C - -Tube 2: C + -Tube 3: TD1 -Tube 4: TD2 -Tube 5: TD5 -Tube 6: TD6 -Tube 7: RBS -Tube 8: ADN -Tube 9: Eprc -Tube 10: Epr1 -Tube 11: MB -Tube 12: KR -Tube 13: Gfp
Sur centrifugeuse à 13 000 Rpm, puis sur prélève le surnageant et sur le remplissage des tubes et appendorf avec les mêmes compositions. On centrifuge une deuxième fois puis sur prélève à nouveau le surnageant. Une fois le double culot de bactéries débarassé de tout surnageant, sur ajoute 250μL de tampon P1, en haut et en bas pour re-suspendre le culot. Attention sur ne racle pas les bactéries. Ensuite sur ajoute 250μL de Buffer P2. (Pas de up and down pour mélanger ici pour ne pas endommager l'ADN génomique et permettre une bonne extraction de celui-ci) et sur ne dépasse surtout pas les 5min dans ce tampon. (Sur remarque la coloration bleu). Idem pour buffer les 350 μL de Buffer N3 ajouté ensuite. (La coloration bleu disparait et on voit apparaitre un précipité blanc. (Les tubes avec P2 puis N3 sont mélangé au principal en inversant le tube).
DAY 7: 14/06/17
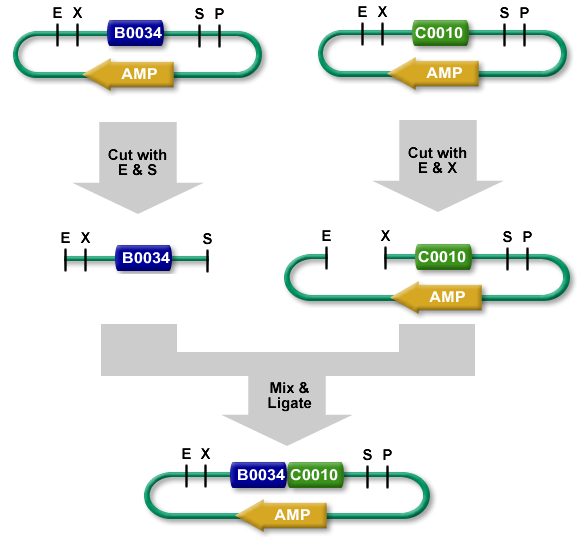
Digestion vector:
We prepared 6 eppendorf tubes, we add in first the buffer ( with the same cone ) and H2O ( with a second cone ). Afterward we add the DNA material for a total mass of 200 ng in each tube. When the initial preparation was done, a short spin was necessary. We extracted the enzyme carefully with a small up action on the edge of the enzymes tubes. Then carefully inject the enzymes in the eppendorf tubes then mix with an up down action. Then we incubate for half an hour at 37° degrees celsius. Afterward, a heat shock is applied at 80°C for 20 minutes to inactivate all enzyme activity.
Tableau / code
Digestion plasmid:
We had 4 tubes of plasmid with each a specific antibiotic resistant gene insert. With a total of volume of 45µL in each. We added 10µL of NE buffer, 41µL of H2O at the end we added our enzymes. ( 2µL for ECORI HF and 2µL for PST1 ). Then we incubate for half an hour at 37° degrees celsius. Afterward, a heat shock is applied at 80°C for 20 minutes to inactivate all enzyme activity.
| Volume | ECORI HF | SPE1 | XBA1 | PST1 | Buffer | H2O | |
| M13 cut up | 3 | 0,5 | 0,5 | / | / | 2 | 14 |
| EPRI cut up | 3,3 | 0,5 | 0,5 | / | / | 2 | 13,7 |
| EPRC cut up | 4,5 | 0,5 | 0,5 | / | / | 2 | 12,5 |
| EPRC cut down | 4,5 | / | / | 0,5 | 0,5 | 2 | 12,5 |
| GFP cut down | 2,2 | / | / | 0,5 | 0,5 | 2 | 14,8 |
| RBS cut down | 1,1 | / | / | 0,5 | 0,5 | 2 | 15,9 |
Ligation:
We prepared 3 tubes for ligation for which we added 2 µL of T4 DNA ligase buffer 10x. Then 11µL of ddH2O, afterward the DNA material ( 2µL upstream 2µL downstream 2µL destination plasmid ). Finally we add T4 DNA ligase and let it incubate at room temperature for 10mn. When the ligation was done, we transformed 5µL of ligation product into 150µL of bacterial cell. ( Previously mentioned ).
Electrophoresis:
As mentioned previously, for each ligation product we added in an eppendorf tube 5µL of DNA 10µL of H2O, 3µL of loading purple dye. In the gel, the order was EPRI cut up, EPRC cut up, EPRC cut down, DNA ladder, RBS cut down, GFP cut down, M13 cut up. We let it migrate for than 30 minutes, at 135 volt which was the limit of the migration, we should have put it for only 25. All fragment under 200 Pb weren't displayed because it had surpassed the migration cap. For EPRI, EPRC cut up and down, we should have seen one bar for the linear plasmid and not for the digested factors.
PHOTO ELECTRO
We also re-did a transformation for RBS which failed previously. This time we used new competent cells and we had a positive result because there were multiple colonies on the gel.
DAY 8: 15/06/17
Préparation of DNA and oligonucleotids:
We recieved 6 tubes of DNA. (P3RsS, p3VcC, p3VcF, p3VcV, p3Xc and p3Xfa). All these tubes are at 1000ng. To be able to prepare them at 50ng / ul we followed the iDT protocol. Centrifuge the tubes 2 min at 3000rpm Addition of 20ul of H20 Vortex Incubation of 20 minutes at 50 ° C Vortex Short spin centrifugation DNA is stored in the freezer -20 ° C
For oligonucleotides: We received the 4 oligonucleotides. To prepare them we followed the recommendations of the producer (Eurogentec). Centrifugation 2 min at 3000rpm Addition of Water to reach a concentration of 100uM Transformation protocol as mentioned before for TD4, Cas9 and dCas9 with the new competent cells prepared on 13/06.
Preparation of competent bacterial cells protocol as mentioned before was done for 100mL of bacterial cells at a OD of 0.478
Resuspension of genomic material from the iGEM plates #4 at the positions as mentioned : 2B, 2D, 2F, 2H, 2G, 2L, 4B, 4D, 4F, 6B, 6D, 6F, 6H, 8B, 8D, 10B, 12B, 12D, 14B This genomic material is a plasmid that codes for a RFP protein with different antibiotic resistances.
A transformation protocol to test competency of bacterial cells was done for the ligation product that was done yesterday. This test was done with the competent bacterial cells from 13/06 and supercompetent cells for a different lab and normal competent cells from another lab. To test the transformation capability we used GFP from our miniprep for a volume of 2 µL.
All transformations were spread out on petri dishes with the specific antibiotic.
We recieved
DAY 9: 16/06/17
Casting of petri dishes (10 amp / 10 kana / 20 chloramphenicol)
PCR checking: Colonies from RBS, TD4, 2B, 2D, 2F, 2H, 2G, 2L, 4B, 4D, 4F, 6B, 6D, 6F, 6H, 8B, 8D, 10B, 12B, 12D and 14B. Box of 15/06 except RBS 14/06. We forgot to put the antibiotics immediately but added them few hours later.
Transformation: Dna from the kit (cas9, 8B and 14B)
Digestion-ligation: The results being negative for the first attempt, we resumed the digestion-ligation of 14/06.
SLIC: We must insert the iDT sequences into the plasmids, which is why it was decided to make a SLIC so that these fragments are inserted into a plasmid with resistance to chloramphenicol (pSB1C3). The DNA selected for SLIC is p3VcV, p3VcF, p3Xc, p3RsS, p3Xfa and pVcC. The ratio used is 3 insert for 1 vector. The number of inserts was calculated using the NEBioCalculator software. The DNA was then transformed into DH5α cells.
DAY 10: 17/06/17
The PCR done on the 6/16 wasn't used because the plugs were not closed. For that reason, our tubes were dry. Despite a rehydratation Malgré une tentative de réhydratation, nous n'avons pas pu récupérer des résultats satisfaisants lors de notre électrophorèse. Nous avons donc re effectué notre PCR. En plus de cela, nos repiquages sur boîtes n'étaient pas vérifiable pour le 6F ( Kanamycine ). -> Repiquage de 6F en Kana. Ensuite : Incubation 37° overnight.
Infection par phage:
-100mL de bactérie Ecoli F+ de la préculture overnight, et ajout de 1µL de phage à 10^5 pfu/µL
-Incubation 50mn ( au lieu de 20 ) à room temperature.
-Depose sur boite de 100µL sur boîte LB agar sans antibio. Témoin bactérie Ecoli F+ sans phage.
-Incubation overnight à 37°
DAY 10: 18/06/17
La PCR réalisé le 6/16/2017 n'a pas pu servir car les bouchons n'avaient pas été fermés dans la machine. Nous avions donc des échantillons sec. Malgré une tentative de réhydratation, nous n'avons pas pu récupérer des résultats satisfaisants lors de notre électrophorèse. Nous avons donc re effectué notre PCR. En plus de cela, nos repiquages sur boîtes n'étaient pas vérifiable pour le 6F ( Kanamycine ). -> Repiquage de 6F en Kana. Ensuite : Incubation 37° overnight.
Infection par phage:
-100mL de bactérie Ecoli F+ de la préculture overnight, et ajout de 1µL de phage à 10^5 pfu/µL
-Incubation 50mn ( au lieu de 20 ) à room temperature.
-Depose sur boite de 100µL sur boîte LB agar sans antibio. Témoin bactérie Ecoli F+ sans phage.
-Incubation overnight à 37°
DAY 11: 19/06/17
RBS, TD4, 2B, 2D, 2F, 2H, 2G, 2L, 4B, 4D, 4F, 6B, 6D, 6F, 6H, 8B, 8D, 10B, 12B, 12D and 14B miniprep. Verification PCR to verify the 3 ligations on Kana that did not give colonies, to know if it was ligation or transformation that was not effective. Electrophoresis.
A) Preparation of the DNA: We received 3 tubes. (P3Xfu, MultiTag and Xpr). All these tubes are at 1000ng. To be able to prepare them at 50ng / ul we followed the iDT protocol. The DNA is stored in the freezer -20 ° C.
B) Infection by phage: Since the infection of 17/06 did not work, we start again with a concentration 1000 times higher. 100 mL of Ecoli F + bacteria from the preculture, and addition of 1 μL phage to 10 ^ 8 pfu / μL
C) Transformation of phage DNA Since the infection of 17/06 did not work, we check that it is not DNA. Transformation of 0.5 μl of phage DNA (10 μl pfu / ml) into 50 μL of DH5α.
DAY 12 : 20/06/17
Amplification protocol for cas9 dcas9 8B and 14B. We were running out of DNA material for these 4. So an amplification was doe. We added in 4 tubes, 50 µL of reaction compounds, composed of Q5 High fidelity 2X Master mix, forward and reverse primer, template DNA and ddH2O. The cycling routine for PCR was 98°C for initial denaturation, then 30 cycles at 98°C, 60, 72, afterward a final extension for 2mn at 72°C. Then hold at 6°C. We verified our PCR, with an electrophoresis. We added 5µL of DNA template. 10µL of H2O, 3µL of loading purple dye. The electrophoresis was set for 25 minutes at 135 volts. The gel was put in gel red for 10 minutes, then revealed using UV. The result are shown as is:
PHOTO
For cas9 / dcas9 we had a negative result, no bounds at 4000 to 5000 pb. And a positive result for 8B and 14B a distinct bound at a thousand pb. A PCR clean up was then done to recuperate the DNA material. We followed the recommendation of the supplier. To our 45µL of DNA, we added 90µL of NT1 buffer. We placed the solution on the column then added 700µL of NT1 buffer. 30 seconds centri at 11 000g. Then 700µL of NT3, centrifuge then discard. A 1mn centri to dry out the column. To recuperate the DNA, 20µL of ddH2O at 70°C was let to incubate on the column for 5mn. Afterward, 20µL of normal H2O. A centri for 1mn at 30 to 50g then another at 11kg. We repeated two times and kept the flow through. We measured the DNA concentration using nanodrop the values mentioned as is.
Cas9 -> 4,48ng/µL dcas9 -> 4,24ng/µL 8B -> 9,54ng/µL 14B -> 11,88ng/µL
We also did a PCR for Cas9 and dCas9.
We have been trying to PCR this two several time in order to be able to use them for mini-prep.
( See PCR protocol ).
Once the PCR was done, we injected our samples on the gel and began electrophoresis.
Once the electrophoresis was done we putted the gel in red for 15mn and then studied it under UV.
Unfortunately no result could be observed on the gel. We decide to stop trying to use Cas9 and dCas9 for the moment.
DAY 13 : 21/06/17
Lisa and jeremy made 19 petri dishes with Kanamycin.
Transplantation of 4 plates of phage from 20/01 in a Kanamicin Petri Dish.
The competence test reveled that our super competent bacteria are approximately three times more competent than Sandra's one, and approximately 1,5 times more competent than Le test de compétence a révélé que nos bactéries super compétente sont en moyenne 3 fois plus compétentes que les compétentes de Sandra, et environ 1.5 fois plus compétentes que les super compétentes de l’équipe voisine.
Le test de fonctionnement de la ligase montre que la ligase n’est pas efficace, et a perdu son activité. Cela est très probablement dû à une mauvaise conservation de celle-ci et aux nombreux chocs thermique qu’elle a subis.
Then we made new competent bacteria. From the overnight bacteria, we made new competent bacteria. Our first dilution resulted in a OD of 0.54 ( 5.4 because the DO was post dilution ). We then added our culture in 200µL of LB in order to get a OD of 0.1. To do that we had to dilute 1/50: Solution: -200mL of LB -4mL of culture. The OD measured post 1h incubation was 0.7 The rest was the same as in the protocol. Putted in Petri dish overnight.
We had to check RBS because of last night. RBS vérification by PCR. ( RBS used : 681 ng/µL ) Height awaited ~ 3000 pd on mini prep (0.2 µL of DNA ). The results from PCR were putted in an electrophoresis and revealed that we failed. 5 stripes when we awaited only 1 ). We remade digestion because we had suspicious results on our electrophoresis. Remaking digestions M13 Making digestion of “red” vector ( RFP ) in order to get linearized plasmids cutted at EP extremity.
Check digestion in gel of EprC / EprI. Digestion of RFP vectors - Purify on agarose gel - CmR et KmR (respectively C3 and K3 - Digest maximum quantity of vectors ~1000ng with 2x1µL of enzymes dans un volume total de 50µl. Let it migrate a long time (40/45 min) to have a good separation. Une fois que la migration est terminée, on révèle au gel red et aux UV. En respectant les règle de sécurités, nous avons à l’aide d’une lame découpé les bandes gel contenant les plasmides linéaires, que nous avons mis dans des tubes eppendorf. Après avoir pesé nos tubes, pour connaître la masse de gel dans chacun de ceux-ci (K3: 0.4g ; C3: 0.3g), nous avons effectué un gel clean-up en suivant le protocol. A l’issue du gel clean-up, nous avons récupéré 60µl de solution de plasmide linéaire, très peu concentré ( environ 2-5 ng/µl).
We made an amplification of Cas9 dCas9 with Pfu turbo which was long graciously with Eric Durand. This protocol consisted of adding 1mL of our DNA template, to 2.5 mL of primers and 1mL of Dntp 5mL of DMSO and 5mL of PFU mix and to add a sufficient quantity of H2O to complete to 50mL. The program for the PCR was set by Eric: 70°C for lengthening, 68° for denaturation, and 58° for oligomerization. The PCR thermocycling took 4h and 44mn.
We did the digestion of 50 ng of iDT DNA sequences p3 VCV, p3 VCC, p3VcF, p3Xfa ,p3Xfu, p3RsS, p3Xc, MultiTag, Xpr in 15µL. We ligate those fragment in pSB1C3 and we did the transformation in DH5Alpha. For the ligation we put 6µL of insert and 2µL of plasmid with 1µL of T4 DNA ligase Then we made an amplification PCR of idt sequences (same one). We used 10ng of DNA for the amplification.
On the petri dish with GM1 E.coli infected with M13 phage, coming from the 20/06, we seen multiple infection site. Thus we transferred 4 of them to a kanamycin petri dish.
Day 14 : 22/06
The M13 infected E.coli Has grown so we pick some colonies in order to make 4 starters. Some of the digestion and ligation of iDT sequence work (p3 VCC, p3 VcV and Xpr) so we make starters. In the temoin plane none colonies were visible.
Day 15 : 23/06
Transplanting GFP, killer red, EprC, EprI, M13. Preparation of starter. Jérémy makes 18 petri dishes Chloram
We received the iDT sequence of p3_Ng and p3_RSM so we eluate them in water (20µL) at the concentration of 5ng/µL.
Camille and Thibault used the starters from MG1 cultures with M13KO7 phages in it. Then, we did mini preps with 4 extracts from different spots on the petri box. GR for “grande”, TP for “tres petit” p1 and p2.
after the minipreps, we measured DNA concentration with nanodrops, our results were : tp :248 ng/µL, GR : 51 ng/µL p1 : 48 ng/µL p2 : 154 ng/µL RBS : 338 ng/µL
In order to verify that we purify M13KO7 genome we digest the phagemid with BamHI and PstI-HF. We digest 100ng of plasmid in 20µL. Thus we observed 2 bands one at 5859 pb and the other at 2830 pb.
With theses concentrations, we made a electrophoresis gel, here are the results :
From left to right :
1 - 2 - 3 - Ladder - Eprc up E/S - M13 up ES - GFP down XP - Epr1 down XP - KillerRed down XP - ladder - 8B - 14B
Digestion:
Vector RFP Ampicillin resistant in order to pick up a linear vector digested on one side by EcorI and on the other side by PftI. The goal is to be able to construct afterward. To do so, we took RFP vector including a gene resistant to ampicillin. We prepared 100µL -> 6µL de plasmide RFP Amp R ( to get approximately 2000 ng ) -> 2µL EcoRI HF -> 2µL Pst I -> 80 H2O -> 10µL of buffer 2.1
30 min 37°C then 20 min 80°C.
We made it migrate on an 1% agarose gel. The gel must be long enough to allow a good separation to see a good split between stripes. Then we revealed it with the usual gel RED+UV protocol. As our stripes were revealed, we cut the corresponding stripes and transfer them in two separate eppendorf tubes. The two tubes weight were 280 and 120 mg respectively. Then we followed the gel clean up protocol : we add 200µL of NT1 to 100 mg of gel, the tube was heated and vortex to dilute the gel in the liquid, after liquification of the gel we poured the solution on a column and let it soak of 5 minute a centrifuge at 10000 g was done to flow through the mix and afterward discarded. 700µL of NT3 was then added, a centrifuge was put in use to pass through NT3 and discarded also. To dry out the column a 1 minute centrifuge was done. To recuperate the DNA we let the column soak for 5 minute with hot water. We eluted the DNA with 30µL of water after centrifuge at 10000g the flow through was kept in a newly added 1.5 mL eppendorf tube. We did the same thing a second time with 20µL of hot water. We measured the DNA concentration using a nanodrop. The concentration were 10 and 12 ng/µL respectively. We finished with an electrophoresis to verify if we actually recuperated the DNA, the extra were kept at -20 degrees. digestion we digested KR, EprC, M13, EprI, and GFP EprC and M13 were digested using the enzymes EcoRi and SpeI KR, EprI and GFP were digested using XbaI and PstI 500 ng of DNA template were digested following the protocol to the letter. to verify an electrophoresis was done, the results are shown and described as follow.
For the binding : Up stream : EprC ; M13 ( EcoRI and Spe I ) Down stream KR ; EprI ; GFP ( XbaI and PstI )
EprC -> 11µL to get 500ng ( 33µL of H2O ) cutsmart M13 -> 7,6µL to get 500ng ( 36,5 of H2O )
KR -> 3,8µL to get 500ng ( 40µL of H2O ) EprI -> 8,5 µL to get 500ng ( 35,5µL of H2O ) GFP -> 5,7 µL to get 500ng ( 38,5 µL of H2O ).
Day 16
Camille and Thibault made minipreps and nanodrops of : Xpr : 4ng/mL -- p3VcV 8ng/mL -- M13 15ng/mL -- EprI 14ng/mL -- GFP 25ng/mL -- KR 57ng/mL -- EprC 18ng/mL.
Jérémy makes 43 competent cell DH5Alpha
Camille did the digestion and ligation of p3Ng and p3RsM in pSB1C3 at a concentration of 10uM. She transform those fragment in DH5a (2µL of dna for 50µL of competent cell). Other ligation where transform by Thibault (6µL of dna for 50µL of competent cell), the fragment were : p3Xc p3Xfu, p3Xfa, MultiT, p3RsS, p3VcF.
Quick changes were made. We wanted to put a mutation on M13KO7 in order to insert a punctual mutation. Thus this mutation will permit the apparition of a restriction site. The Quick Change is a PCR ON. We have two slot : 1 → M13KO7Tp and 2 → M13KO7p2. Firstly, we used the primers Bdw and Bup to insert the BspI.
Quick change : Plasmid : 1µL dNTP(10mM) : 6µL DMSO : 1µL Primer 1 : 2,5 µL Primer 2 : 2,5 µL Tampon pfu turbo : 5 µL pfu Turbo : 1 µL H2O qsp 50 µL
I put primers with a concentration of 100mM. This might be to concentrate….