Erboardman (Talk | contribs) |
|||
| Line 338: | Line 338: | ||
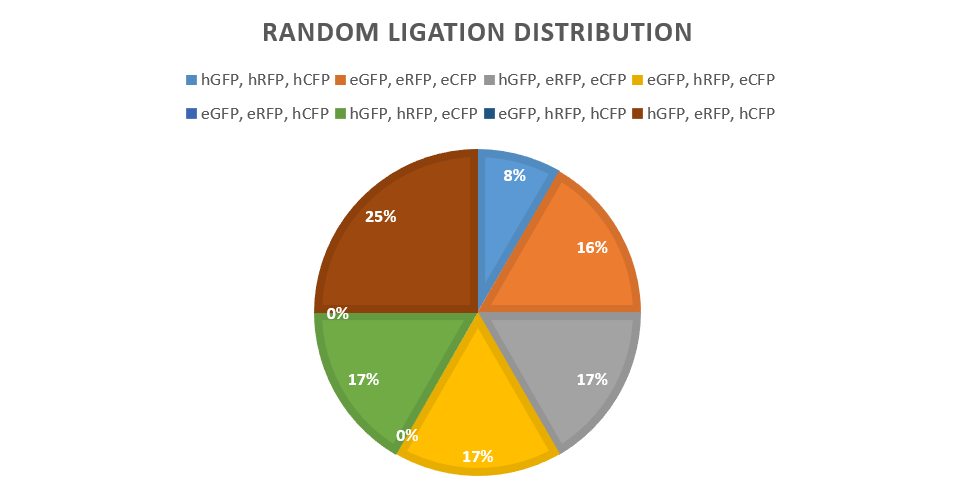
<p> The highest and lowest performing promoters were chosen to give the most easily visible result. Promoter E and Promoter 4. Promoter 4 gives a high expression of fluorescent proteins, as shown by our promoter library findings. The promoters were then attached to each reporter protein CFP, RFP and GFP via BSAI digestion and ligation (creating no scar sites) along with a terminator to form six "brick" variants. After amplification of the bricks produced via PCR, seven products combinations were ligated to a low copy backbone in the pattern of red, green and then blue (fluorescent protein) consistently through controlled use of the DCBA digestion site. In addition to a set of "random ligations". Each ligation has only one ligation slot (due to availability of restriction cut sites) per reporter type/colour, leaving random chance to produce a combination of all the possible variants in the random ligations where multiple promoters are available.</p> | <p> The highest and lowest performing promoters were chosen to give the most easily visible result. Promoter E and Promoter 4. Promoter 4 gives a high expression of fluorescent proteins, as shown by our promoter library findings. The promoters were then attached to each reporter protein CFP, RFP and GFP via BSAI digestion and ligation (creating no scar sites) along with a terminator to form six "brick" variants. After amplification of the bricks produced via PCR, seven products combinations were ligated to a low copy backbone in the pattern of red, green and then blue (fluorescent protein) consistently through controlled use of the DCBA digestion site. In addition to a set of "random ligations". Each ligation has only one ligation slot (due to availability of restriction cut sites) per reporter type/colour, leaving random chance to produce a combination of all the possible variants in the random ligations where multiple promoters are available.</p> | ||
<p> e= empty promoter, h= high expression promoter</p> | <p> e= empty promoter, h= high expression promoter</p> | ||
| + | <p><span style="color: #ffffff;"> </span></p> | ||
| + | <p><span style="color: #ffffff;"> </span></p> | ||
<center><img src="https://static.igem.org/mediawiki/2017/a/a1/T--UNOTT--brickstitching.jpeg"></center> | <center><img src="https://static.igem.org/mediawiki/2017/a/a1/T--UNOTT--brickstitching.jpeg"></center> | ||
| + | <p><span style="color: #ffffff;"> </span></p> | ||
| + | <p><span style="color: #ffffff;"> </span></p> | ||
<p> These bricks were ligated with the backbone as follows: </p> | <p> These bricks were ligated with the backbone as follows: </p> | ||
<li> eRFP-eGFP-eCFP - Low expression in all reporters</li> | <li> eRFP-eGFP-eCFP - Low expression in all reporters</li> | ||
| Line 346: | Line 350: | ||
<li> eRFP-eGFP-hCFP - Low expression in RFP and GFP, but High expression of CFP</li> | <li> eRFP-eGFP-hCFP - Low expression in RFP and GFP, but High expression of CFP</li> | ||
<li> eRFP or hRFP-eGFP or hGFP-eCFP or hCFP - As both variants of each reporter brick available, every combination is possible. </li> | <li> eRFP or hRFP-eGFP or hGFP-eCFP or hCFP - As both variants of each reporter brick available, every combination is possible. </li> | ||
| + | <p><span style="color: #ffffff;"> </span></p> | ||
| + | <p><span style="color: #ffffff;"> </span></p> | ||
<p> This produced a control for each reporter fluorescent protein in isolation. It also produced a control for all reporters on either highest expression or lowest expression. This finally also produced a random set of colonies for isolation (after transformation) for comparison and test for true randomness.</p> | <p> This produced a control for each reporter fluorescent protein in isolation. It also produced a control for all reporters on either highest expression or lowest expression. This finally also produced a random set of colonies for isolation (after transformation) for comparison and test for true randomness.</p> | ||
<h1>Results</h1> | <h1>Results</h1> | ||
Revision as of 00:28, 2 November 2017

EXPERIMENTS: