MattAFrench (Talk | contribs) |
MattAFrench (Talk | contribs) |
||
| Line 319: | Line 319: | ||
<h1>Results</h1> | <h1>Results</h1> | ||
| − | <p><img src="https://static.igem.org/mediawiki/2017/3/30/T--UNOTT--Promoterpoolstrength.png"> | + | <p><table> |
| − | <img src="https://static.igem.org/mediawiki/2017/3/30/T--UNOTT--Promoterpoolstrength.png"><br>Figure 1 illustrates the strengths of each set of strong (SP) and weak (WP) RBS promoters, the first graph being the SPs 1, 2, 3, 4, 5, and E. E is a null promoter which will produce no expression, providing an additional control like sgRNA0 is for the gRNA parts design. It will help making sure no unspecific interference happens when no promoter is present. The second graph shows W1-4. It is clear there are some extremely different expression levels. SP3 and SP4 are quite similar with peaks of 5500-5800 a.u. at 24 hours; it would be hard to discern between them, but these are significantly the strongest promoters and would be useful as a ‘maximum’ expression promoter. SP1 and W4 are also quite comparable with peak around 3000 a.u., these would be considered intermediate/strong promoters. W3, W1, SP2 and perhaps SP5 are all relatively similar with peaks from 900-1700 a.u. at 24 hours. These would be considered weak promoters, they would be useful in a triple reporter experiment to act as the separate targets for the sgRNAs and see if they have distinct inhibited states. W2 is by far the weakest active promoter, with a peak at >500 it would be classed as the weakest promoter, perhaps serving as a minimum activation promoter. SE as expected produced no expression, and can be concluded it will work as a negative control or null promoter. | + | <tr><img src="https://static.igem.org/mediawiki/2017/3/30/T--UNOTT--Promoterpoolstrength.png"></tr> |
| + | <tr><img src="https://static.igem.org/mediawiki/2017/3/30/T--UNOTT--Promoterpoolstrength.png"></tr> | ||
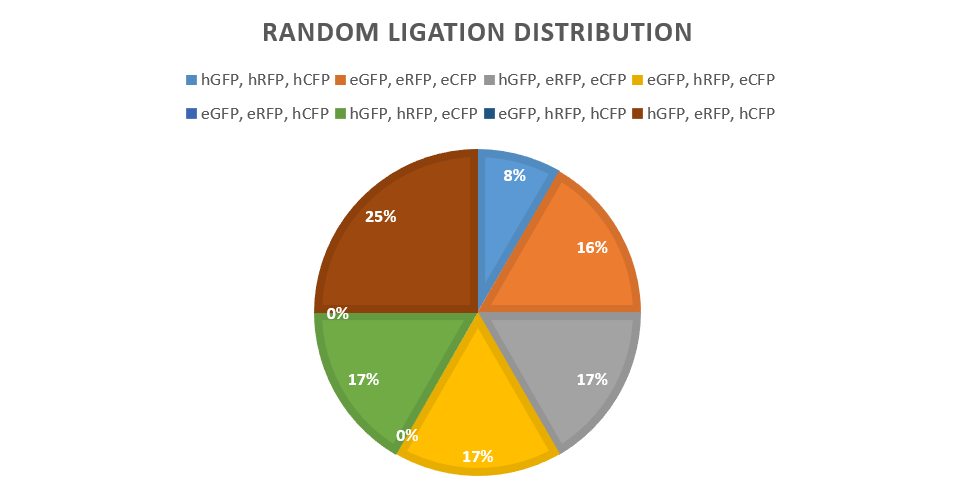
| + | <tr><th>Figure 1. This graph shows the relative fluorescent intesnity of strong and weak RBS promoters SP1-E and WP1-4. These were grown for a 24 hour period and measurements were taken at 0, 2, 4, 6, 9, and 24 hours.</th></tr> | ||
| + | <br>Figure 1 illustrates the strengths of each set of strong (SP) and weak (WP) RBS promoters, the first graph being the SPs 1, 2, 3, 4, 5, and E. E is a null promoter which will produce no expression, providing an additional control like sgRNA0 is for the gRNA parts design. It will help making sure no unspecific interference happens when no promoter is present. The second graph shows W1-4. It is clear there are some extremely different expression levels. SP3 and SP4 are quite similar with peaks of 5500-5800 a.u. at 24 hours; it would be hard to discern between them, but these are significantly the strongest promoters and would be useful as a ‘maximum’ expression promoter. SP1 and W4 are also quite comparable with peak around 3000 a.u., these would be considered intermediate/strong promoters. W3, W1, SP2 and perhaps SP5 are all relatively similar with peaks from 900-1700 a.u. at 24 hours. These would be considered weak promoters, they would be useful in a triple reporter experiment to act as the separate targets for the sgRNAs and see if they have distinct inhibited states. W2 is by far the weakest active promoter, with a peak at >500 it would be classed as the weakest promoter, perhaps serving as a minimum activation promoter. SE as expected produced no expression, and can be concluded it will work as a negative control or null promoter. | ||
In conclusion, it is promising that we have 5 distinct states of expression with maximal, strong, weak, minimal, and off. There is also the opportunity to have SP2, W1, W2 and maybe SP5 to act as a way to discern how effective the sgRNA:dCas9 inhibition, owing to their similar expression patterns. | In conclusion, it is promising that we have 5 distinct states of expression with maximal, strong, weak, minimal, and off. There is also the opportunity to have SP2, W1, W2 and maybe SP5 to act as a way to discern how effective the sgRNA:dCas9 inhibition, owing to their similar expression patterns. | ||
</p> | </p> | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
Revision as of 02:27, 2 November 2017