
Results
Over the summer, we went through a lot of experiments and generated many results. The ones central to our project are analyzed below. Before we could start experimenting with triggers and toeholds however, we had to learn a few things about our cell-free reagents and how they are made. For this we got familiar with making lysates and energy solution for cell-free extracts, as can be seen here. Once this basis was (somewhat) established, we were able to go on with trigger/toehold reactions.
- Testing RNA trigger with toehold in PURExpress, RNA titration.
- Testing RNA trigger with toehold in lysate.
- Successful α-complementation.
- Testing toehold lacZalpha in lysate.
- Testing short single stranded DNA triggers in lysate, short ssDNA titration
- Testing short single stranded DNA triggers in lysate, long ssDNA titration
- Testing aptamer trigger in PURE, aptamer trigger titration in PURExpress.
- Testing aptamer trigger in lysate, aptamer trigger titration in lysate.
- Successfully generating toeholds for Hepatitis C and testing them in our lysate.
What was accomplished ?
In order to chronologically summarize our best accomplishments and discoveries during the elaboration of our project before presenting our results, here is a list:
In vitro transcription of trigger
In order to perform a toehold reaction, we first had to make the trigger. To simulate the environment of viral RNA within a sample, an in vitro transcription was necessary to get the trigger as an RNA molecule. We consequently purified the RNA trigger by LiCl2 precipitation (see this protocol). The transcription product (both purified and unpurified) was then analyzed on an Urea-PAGE gel.

We see the expected band of ~390 base pairs on the gel and can conclude that we produced RNA trigger. The bands of the purified trigger are a lot fainter, as expected.
Testing RNA trigger in PURE
Next we went on to test whether the transcribed RNA sequence would trigger the toehold. For this we used in a first experiment a commercially available cell-free synthesis system (PURExpress, further simply called PURE). The toehold was taken from the paper by Pardee et al1, number 27 from the B series. Reaction components were added as listed in the table below and measured on a platereader at 37°C.

Measuring absorbance at 595 nm

The toehold was triggered by the RNA trigger and beta-galactosidase expressed, the solution in the tube changed colour from yellow to purple.
Testing RNA trigger in lysate
Upon those first encouraging results we decided to test the RNA trigger in our own T7 M15 cell lysates to assess whether the switch works in our cell-free expression system.
We incubated at 37°C the reactions listed in the tables below and recorded the results after 1 hour of incubation:



In our self-made lysate the toehold was triggered by the RNA trigger and beta-galactosidase expressed, the solution in the tube changed colour from yellow to purple. This was a big result for us as it meant that we had successfully repeated Pardee's1 experiment!
Titration of RNA trigger
After successfully triggering the toehold in our cell-free lysates, we were interested in the limit of detection of the RNA in our reaction i.e the minimal quantity of RNA that would give us a colorimetric output detectable by eye. To do so, we performed a titration of the in vitro transcribed RNA in PURE.


To summarize this result, we created the following titration curve :

After assessing that the limit of detection was about 720 nm of RNA in this experiment, we were interested in exploring this result and extrapolating it to different nucleic triggers.
Alpha complementation
To get started, we first tried to prove that we can obtain a functional beta-galactosidase starting from a T7 lacZ alpha DNA template added to the T7 M15 lysate reaction.


Testing LacZalpha toehold
After proving that alpha complementation worked in our cell-free platform, we went on to test this concept for LacZ alpha contained in a toehold switch.

Right : Toehold LacZ alpha with no trigger added: the alpha part cannot be translated
We can see that the solution turned purple in the tube where trigger was added. This means that a complete beta-galactosidase was present, indicating that the toehold unfolded and LacZ alpha was translated.
Testing ssDNA triggers in lysate
Planning to use the toehold as signal relayer / signal transducer when an aptamer pair detects a target protein, we wanted to have an idea on how a single stranded DNA would interact with the toehold and quantify the detection limits before exploring aptamer-toehold interactions.
To do so, we first tried to demonstrate that the toehold could be triggered by single stranded DNA triggers of different lengths: a 36 base-pair single stranded DNA (ssDNA short) which is exactly complementary to a sequence within the toehold and a 90 base-pair single stranded DNA (ssDNA long) which has flanking bases upstream and downstream of the complementary toehold sequence.



Titration of ssDNA short trigger in lysate
After showing that the single stranded DNA sequences triggered the toehold we wanted to quantify the signal depending on the concentration of single stranded DNA present in the sample.
We performed titrations of the single stranded DNA triggers in our own cell lysates of T7M15 to assess the limit of detection of our system.


The signal was clearly different from the negative control for all of the concentrations we tested. This implied that a concentration as little as 500nM is sufficient to trigger the toehold switch and produce the colour change.
Titration of ssDNA long trigger in lysate
The same titration experiment was performed to find the lowest trigger concentration that could still achieve expression of beta-galactosidase.


The results showed that the limit of detection of the ssDNA long trigger lies at 2.5 µM. It suggested that the flanking bases upstream and downstream of the ssDNA long sequence were interfering with the annealing of the trigger to the toehold. This would then lead to lower sensitivity of the toehold in presence of a trigger.
This presented a first insight into the fact that working with aptamers in order to trigger the toehold may require higher aptamer concentrations, because of the 3D conformation of the aptamer and the extra base pairs upstream of the complementary sequence.
Zika 32B toehold
When the time came to start cloning, we wanted to test our cloning strategy on another Zika toehold from Pardee's paper1. We decided to go for the series B, number 32 toehold.

The 32B toehold expressed beta-galactosidase well and is in line with our results found previously.
Aptamer trigger in PURE
After these encouraging results with using a single stranded DNA sequence as a toehold trigger, we decided to use a ssDNA sequence bound to an aptamer for our next experiment. We added the ssDNA short trigger sequence to one of the aptamers used to detect our protein of interest.
This would give us first insight into how the detection scheme would work as a whole, and whether the concentrations of aptamers needed to detect the target protein would match that of trigger needed to unfold the toehold.
To answer those questions, we first performed an aptamer-trigger titration in PURE.



The results above show that the limit of detection of aptamer trigger in PURE is around 50 nM.
Titration of aptamer trigger in lysate
After those promising results, we performed the titration of the aptamer-trigger in our T7M15 cell lysates to see if we could replicate similar results


The titration of aptamer trigger in T7M15 cell lysates showed that the limit of detection was around 2.5 µM.
As a conclusion : We demonstrated that the toehold can be triggered by single stranded DNA, RNA and aptamer triggers. We also characterized the limit of detection of each trigger form.
Testing the softare : New Hepatitis C toeholds
Using the Toehold Designer, our software, we created four new toeholds for Hepatitis C, genotype 1a. We decided to test our software for this particular virus after having contacted professionals working in the field as part of our Integrated Human Practices.
We performed the usual toehold reaction with those new toeholds : 10 µl reactions, measuring absorbance at 595nm on a plate reader, at 37°C. Those reactions were all set up as follows : 2.5 µl Energy Solution, 2.5 µl Buffer A, 2.5 µl lysate mix (T7 M15 + Top10-GamS), as well as toehold (final concentration 4 nM), trigger (final concentration 1 µM), substrate and water. Absorbance was measured on a plate reader at 595nm and 37°C. Below: the four new toeholds compared and a graph showing each toehold's efficiency separately.

Shaded error graphs

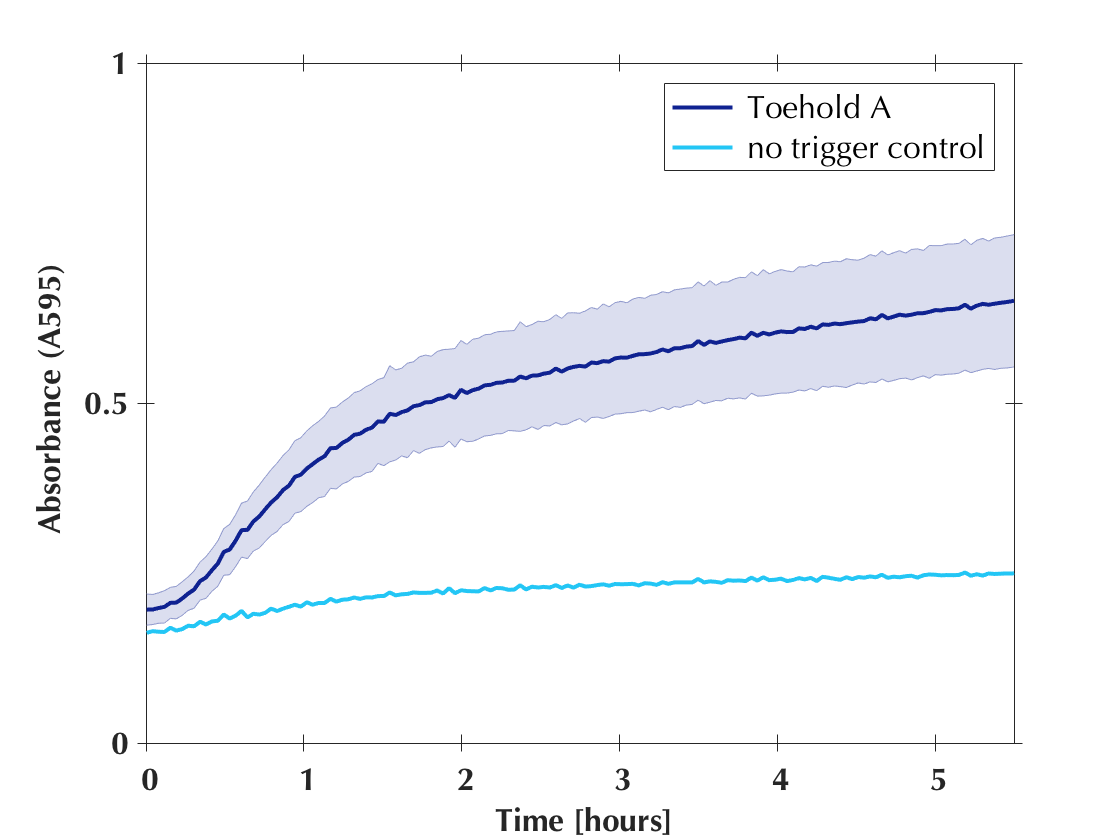



All the toeholds generated by our software worked! A colour change visible by eye was observed for every reaction. This result clearly demonstrated the usefulness of our software and confirmed its efficiency. Shortly thereafter we published the code to our software on GitHub to make it available to anyone who wants to create new toehold switches!
References
1. Pardee, Keith, et al. "Rapid, low-cost detection of Zika virus using programmable biomolecular components." Cell 165.5 (2016): 1255-1266.



