| Line 3: | Line 3: | ||
<html> | <html> | ||
| − | < | + | <script type="text/javascript" src="path-to-MathJax/MathJax.js"></script> |
| − | + | <h1>Modeling</h1> | |
| − | + | ||
| − | <h1> | + | |
<h3>Estimate and optimize yield.</h3> | <h3>Estimate and optimize yield.</h3> | ||
<body align="center"> | <body align="center"> | ||
| − | |||
<div align="left"> | <div align="left"> | ||
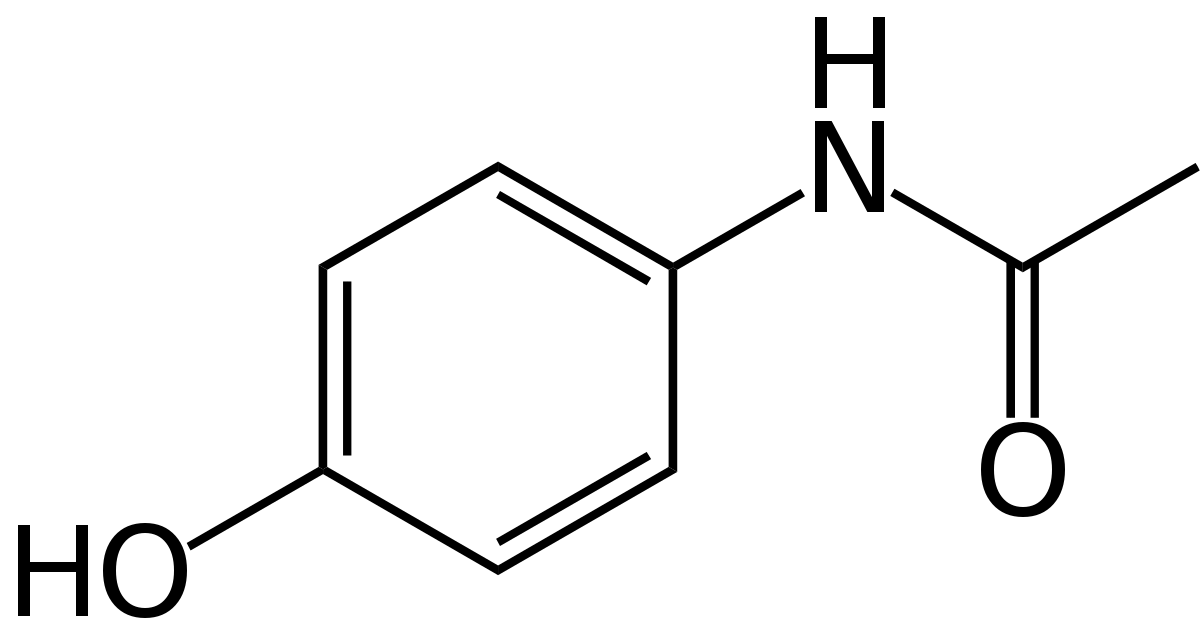
| − | <h4>Acetaminophen</h4> | + | <h4>Acetaminophen <img class="acetaminophen" src="https://upload.wikimedia.org/wikipedia/commons/thumb/2/29/Paracetamol-skeletal.svg/1200px-Paracetamol-skeletal.svg.png" style="width:168px;height=128px"> </h4> |
| + | |||
To predict theoretical acetaminophen production, we calculated the amount of a precursor, chorismate, by quantifying its main products, the aromatic amino acids phenylalanine, tyrosine, and tryptophan. | To predict theoretical acetaminophen production, we calculated the amount of a precursor, chorismate, by quantifying its main products, the aromatic amino acids phenylalanine, tyrosine, and tryptophan. | ||
Since no amino acid composition data was available for Synechococcus, we started by using literature data for similar cyanobacteria species Spirulina and Synechocystis and found that between one and 13.6 percent of amino acids were aromatics by mass, or between XX and YY molar percent. | Since no amino acid composition data was available for Synechococcus, we started by using literature data for similar cyanobacteria species Spirulina and Synechocystis and found that between one and 13.6 percent of amino acids were aromatics by mass, or between XX and YY molar percent. | ||
| Line 24: | Line 22: | ||
</div> | </div> | ||
<div align="left"> | <div align="left"> | ||
| − | <h4>Biomass</h4> | + | <h4>Biomass <img class="spirulina_pic" src="https://static.igem.org/mediawiki/2017/5/5c/Spirulina_powder.png" style="width:168px;height=128px"></h4> |
To understand the production capacity of our organism, we aggregated growth data from published papers and all of our lab’s growth data. Using limited logistic growth curves and linear algebra to fit our equation, we modelled dried biomass and cell count per time, with the additional dependent variables of temperature, light intensity, and starter culture density. | To understand the production capacity of our organism, we aggregated growth data from published papers and all of our lab’s growth data. Using limited logistic growth curves and linear algebra to fit our equation, we modelled dried biomass and cell count per time, with the additional dependent variables of temperature, light intensity, and starter culture density. | ||
<div id="container"></div> | <div id="container"></div> | ||
| Line 126: | Line 124: | ||
background-color: honeydew; | background-color: honeydew; | ||
} | } | ||
| + | img.acetaminophen { | ||
| + | position: relative; | ||
| + | top: 25px; | ||
| + | } | ||
| + | img.spirulina_pic { | ||
| + | position: relative; | ||
| + | top: 40px; | ||
| + | } | ||
| + | |||
</style> | </style> | ||
| Line 200: | Line 207: | ||
title: { | title: { | ||
| − | text: ' | + | text: '' |
}, | }, | ||
subtitle: { | subtitle: { | ||
| − | text: ' | + | text: '' |
}, | }, | ||
yAxis: { | yAxis: { | ||
Revision as of 00:01, 20 September 2017