Raj Magesh (Talk | contribs) |
Raj Magesh (Talk | contribs) |
||
| Line 24: | Line 24: | ||
<h2>Modification — <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2319001">BBa_K2319001</a> (HindIII+ATG+AgeI scar)</h2> | <h2>Modification — <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2319001">BBa_K2319001</a> (HindIII+ATG+AgeI scar)</h2> | ||
| − | <p>A HindIII restriction site, a start codon and an AgeI restriction site (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2319001">BBa_K2319001</a>) are added immediately downstream of the T7 promoter+RBS. A number of design considerations went into this choice of restriction sites</p>. | + | <p>A HindIII restriction site, a start codon and an AgeI restriction site (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2319001">BBa_K2319001</a>) are added immediately downstream of the T7 promoter+RBS. A number of design considerations went into this choice of these restriction sites</p>. The HindIII site — sandwiched between the RBS and the start codon — has the nucleotide sequence 5'-A\AGCTT-3', which is very similar to the optimal sequence predicted by the sequence logo of <i>E. coli</i> <a href="http://parts.igem.org/Help:Ribosome_Binding_Site">ribosome binding sites</a>. |
| + | |||
| + | <img src="https://static.igem.org/mediawiki/parts/d/d1/Ribologo-small.gif"> | ||
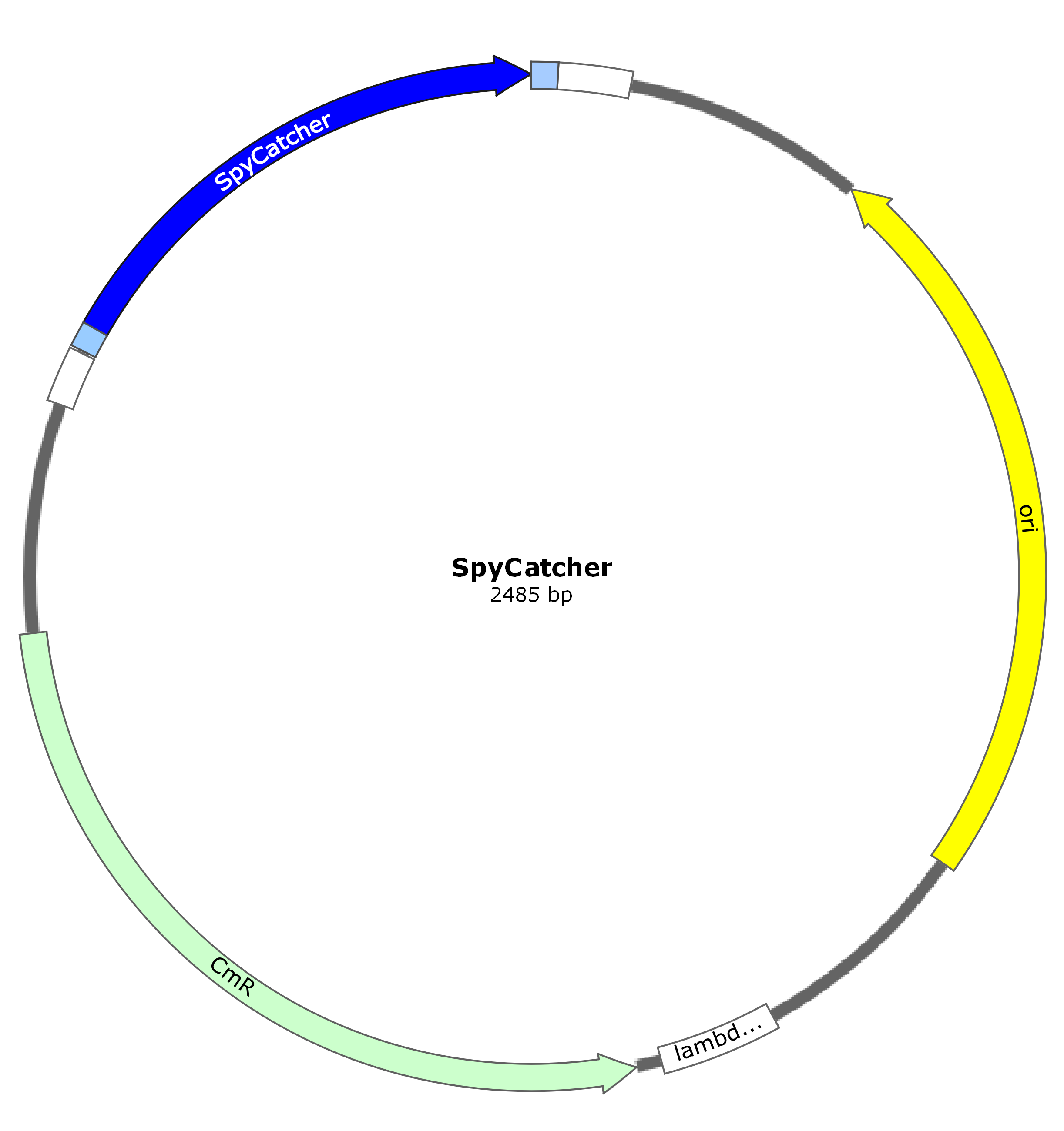
<h1 id="sfgfp-spycatcher">sfGFP-SpyCatcher</h1> | <h1 id="sfgfp-spycatcher">sfGFP-SpyCatcher</h1> | ||
Revision as of 12:47, 30 October 2017
Designing a T7 expression system
Our third method to induce gas vesicle aggregation (SpyCatcher-SpyTag binding) involves protein overexpression, and no system is better than E. coli strain BL21 (DE3)'s T7 expression system for this purpose: BL21 (DE3) is deficient in lon and ompT proteases. BL21 (DE3) has the T7 RNA polymerase gene integrated into its genome under the lac operon; adding IPTG induces expression of T7 RNA polymerase, which recognizes the T7 promoter sequence. Any gene inserted downstream of the T7 promoter can thus be expressed.
Using BBa_K525998 (T7 promoter+RBS) and BBa_K731721 (T7 terminator), we have designed a T7 expression backbone that can be used to assemble and express fusion proteins easily.


Choice of BioBricks
BBa_K525998 (T7 promoter+RBS) was chosen since a strong RBS B0034 is used, and this allows for maximal expression of our proteins of interest. BBa_K731721 (T7 terminator) was chosen instead of the standard B0015 double terminator as its in vivo termination efficiency is greater, as characterized by BBa_K731700.
Modification — BBa_K2319001 (HindIII+ATG+AgeI scar)
A HindIII restriction site, a start codon and an AgeI restriction site (BBa_K2319001) are added immediately downstream of the T7 promoter+RBS. A number of design considerations went into this choice of these restriction sites
. The HindIII site — sandwiched between the RBS and the start codon — has the nucleotide sequence 5'-A\AGCTT-3', which is very similar to the optimal sequence predicted by the sequence logo of E. coli ribosome binding sites.
sfGFP-SpyCatcher

mCherry-SpyTag












