| Line 892: | Line 892: | ||
<br> | <br> | ||
| − | + | /* <b>Starting Density:</b> | |
<button id="MinusDensity" onclick="MinusDensity(); MakeChart();"> < </button> | <button id="MinusDensity" onclick="MinusDensity(); MakeChart();"> < </button> | ||
<var id="htmlDensityID"></var> g biomass/ L | <var id="htmlDensityID"></var> g biomass/ L | ||
| − | <button onclick="PlusDensity(); MakeChart();"> > </button> | + | <button onclick="PlusDensity(); MakeChart();"> > </button>*/ |
</div> | </div> | ||
</div> | </div> | ||
| Line 1,012: | Line 1,012: | ||
document.getElementById("htmlIntensityID").innerHTML = localStorage.jsIntensityValue; | document.getElementById("htmlIntensityID").innerHTML = localStorage.jsIntensityValue; | ||
document.getElementById("htmlTempID").innerHTML = localStorage.jsTempValue; | document.getElementById("htmlTempID").innerHTML = localStorage.jsTempValue; | ||
| − | + | /* document.getElementById("htmlDensityID").innerHTML = localStorage.jsDensityValue; */ | |
function MinusDays() { | function MinusDays() { | ||
| Line 1,031: | Line 1,031: | ||
} | } | ||
function PlusOneLight() { | function PlusOneLight() { | ||
| − | localStorage.jsIntensityValue=parseInt(localStorage.jsIntensityValue)+10; | + | if(localStorage.jsIntensityValue<300){ |
| − | + | localStorage.jsIntensityValue=parseInt(localStorage.jsIntensityValue)+10; | |
| + | } | ||
document.getElementById("htmlIntensityID").innerHTML=localStorage.jsIntensityValue; | document.getElementById("htmlIntensityID").innerHTML=localStorage.jsIntensityValue; | ||
} | } | ||
| Line 1,043: | Line 1,044: | ||
} | } | ||
function PlusOneDegree() { | function PlusOneDegree() { | ||
| − | localStorage.jsTempValue=parseInt(localStorage.jsTempValue)+1; | + | if(localStorage.jsTempValue<37){ |
| + | localStorage.jsTempValue=parseInt(localStorage.jsTempValue)+1; | ||
| + | } | ||
document.getElementById("htmlTempID").innerHTML = localStorage.jsTempValue; | document.getElementById("htmlTempID").innerHTML = localStorage.jsTempValue; | ||
} | } | ||
| − | + | /* function MinusDensity() { | |
if(localStorage.jsDensityValue>0.15){ | if(localStorage.jsDensityValue>0.15){ | ||
localStorage.jsDensityValue=(Number(localStorage.jsDensityValue)-0.1).toFixed(1); | localStorage.jsDensityValue=(Number(localStorage.jsDensityValue)-0.1).toFixed(1); | ||
| Line 1,055: | Line 1,058: | ||
localStorage.jsDensityValue=(Number(localStorage.jsDensityValue)+0.1).toFixed(1); | localStorage.jsDensityValue=(Number(localStorage.jsDensityValue)+0.1).toFixed(1); | ||
document.getElementById("htmlDensityID").innerHTML = localStorage.jsDensityValue; | document.getElementById("htmlDensityID").innerHTML = localStorage.jsDensityValue; | ||
| − | } | + | }*/ |
</script> | </script> | ||
<script src="https://code.highcharts.com/highcharts.src.js"></script> | <script src="https://code.highcharts.com/highcharts.src.js"></script> | ||
Revision as of 14:26, 1 November 2017

MODELING
Predict and optimize yield
Background
The purpose of modeling is to carefully examine the pathways of each intended biosynthetic products, look for ways to optimize production, understand limiting factors, and to support and innovate for the team in wet lab. To accomplish these goals, we read dozens of different academic papers, sorted through metabolic pathways, and used several different methods to model acetaminophen, B12, and biomass production. Each of these modeling methods has different assumptions which allow these data points to be averaged; providing reasonable quantitative estimates of our biosynthetic products.
ACETAMINOPHEN
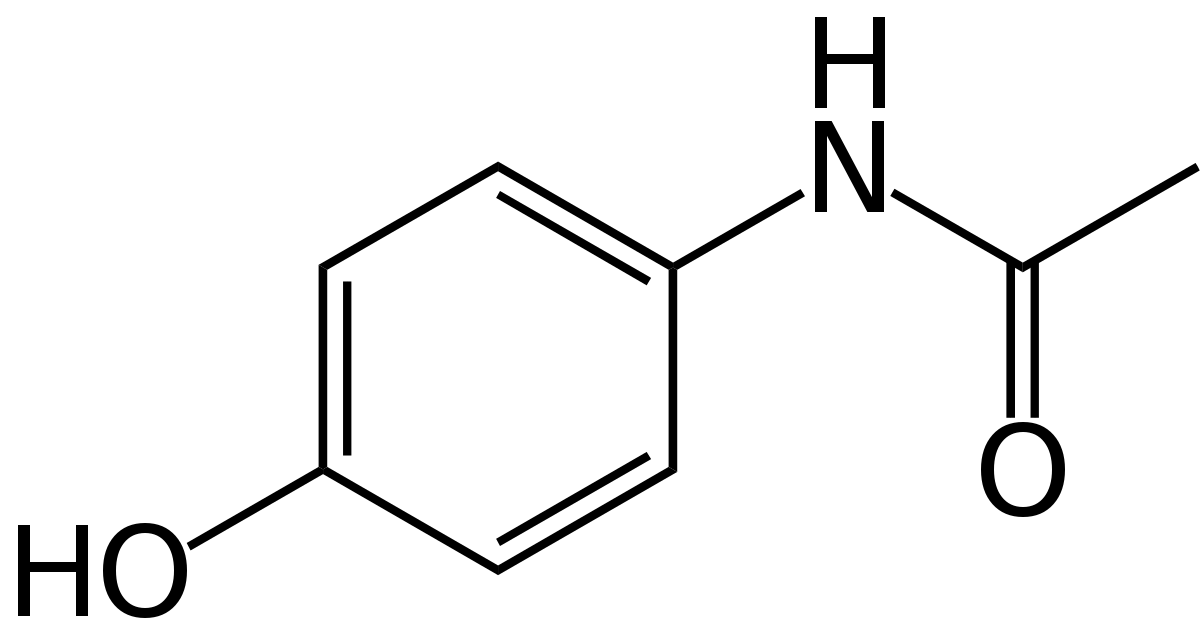
Overview
To predict acetaminophen biosynthesis, we analyzed the abundance of the acetaminophen's precursor, chorismate. Chorismate is a precursor in our organism for the three aromatic amino acids and folate[1]; also producing alkaloids, salicylic acid, and vitamin K in other organisms[2]. Our inserted gene 4ABH takes intermediates from tryptophan and folate pathways and converts it to 4-aminophenol, processing it to acetaminophen with the help of nhoA[3]. We used published data on these intermediates to quantifiy the precursors available for converting to acetaminophen. To understand how much precursor our enzyme 4ABH would process, we compared enzyme Kms against competiting enzymes using ratios and creating a simulation of chorismate metabolism in Python[4]. Insufficient data on enzyme rates, quantities, and Km binding affinity values necessitated using limited data from other bacterial species and comparing sequence identities to find data for the closest sequence to Arthrospira platensis. With these data, we made several estimates for the amount of acetaminophen precursor available and how much would go down each pathway.

Enzyme Competition
Precursor concentrations would be the first limit to how much acetaminophen could be produced. The second would be how effective our enzyme 4ABH was at converting precursors from the tryptophan and folate pathways into 4-aminophenol. To answer the competition question, we can either assume all three competing enzymes will have similar precursor affinity and produce each product in equal quantities or use ratios of each competing enzyme's Km for the limiting intermediates. This enzyme kinetics based method is difficult because there is so little data on rates, quantities, and affinities for our particular enzyme sequences. For that reason, all of the following calulations will be computed using a 33% assumed precursor to product conversion rate.
Since few cyanobacterial proteins have been isolated and tested for kinetic data, many of the Kms compared are from other species. 4ABH has a Km of 20.4 µM for p-amino benzoate(PABA), while the folate processing enzyme folp1 has a Km of 0.37 µM [5,6]. Assuming the rate and quantity are the same, the folate enzyme will be fully saturated at a much lesser concentration of PABA than 4ABH. You can assume that the logrithmic Km ratio of 20.4:0.37 (or 55:1) will represent how many moles of PABA goes to folate versus to acetaminophen. For the tryptophan intermediate anthranilate, no Km is given but 4ABH is assayed as having a rate of 24% of PABA, equal to 34 µM for anthranilate, this compares to TrpD's Km for chorismate of 40nM, meaning we'd have 850:1 tryptophan to acetaminophen. Since these ratios are based on poorly matching enzymes and make acetaminophen production undetectably small, we will keep this enzyme competition in mind while using 33% conversion for simplicity.
Assumptions
- We assume that our genes were designed, inserted, and translated successfully and in reasonable quantities.
- Km values for Arthrospira platensis can be approximated by using other organisms' enzyme rates. Sequences were BLAST compared in each case, resulting in a range of similarity between 52% and 63% positive alignment, suggesting that enzyme orthologs might be significantly different.
- We assume that enzyme rate and quantity is the same for each 4ABH, folp1, and TrpD, else we could not compare Km ratios and calculate approximate the Km of 4ABH for anthranilate.
Amino Acid Method
Once we found out that acetaminophen was produced from the same precursors as the tryptophan and folate, we found published amino acid composition data for Arthrospira platensis and back converted those amino acids to moles of acetaminophen precursors anthranilate and PABA. These molecules are the direct substrate for our enzyme 4ABH, converting it to 4-aminophenol before nHoa converts it to acetaminophen. We show several different calculations below using different sources of data.
Assumptions
- Synechococcus and Arthrospira platensis have similar amino acid ratios. Since there was no available amino acid data for our transformed Synechococcus, we must assume our species has a similar ratio to the more well described Arthrospira platensis.
- The amount of anthranilate and PABA precursors are equal to that of their products tryptophan and folate.
- Of the available precursors, 33% will go down our pathway. This is based off the idea that the enzymes may have similar affinities for the precursor.
- Folate is present in such small amounts, at 0.21 nanomoles of PABA, that its effect on acetaminophen production is neglegible [7].
$$\frac{0.442\ µmol\ Trp}{1g\ biomass}\approx \frac{0.455\ µmol\ anth}{1g\ biomass}\times\frac{1\ mol\ acet}{3\ moles\ anth}\time\frac{151g\ acet.}{1 mol acet.}=\frac{2.3mg\ acet.}{1g\ biomass}$$
Sequence Analysis Method
To validate our organism's quantity of tryptophan precursor, we used a custom Python program[9] to convert DNA sequences to amino acids and calculate molar and mass percentages of tryptophan. We ran both the genome and all 55 listed ribosomal protein sequences through our program, which resulted in 0.9% and 0.6% tryptophan by moles. Knowing that about 60% of Arthrospira platensis is protein by mass, we can predict acetaminophen production.
$$\frac{0.056\ g\ Trp}{1\ biomass}\rightarrow\frac{0.27\ mmol\ Trp}{1\ g\ biomass}\times\frac{1\ mol\ acet}{3\ mol\ Trp}\times\frac{151.163g\ acet}{1\ mol\ acet.}=\frac{13.7mg\ acet.}{1g\ biomass}$$$$\frac{0.036 g\ Trp}{1 g\ biomass}\rightarrow \frac{0.0.175\ mmol\ Trp}{1\ g\ biomass} * \frac{1\ mol\ acet}{3\ mol\ chor} *\frac{151.163\ g}{1 mol\ acet} = \frac{8.8\ mg\ acet}{1 g\ biomass}$$
Assumptions
- Codon composition from the genome and ribosomal proteins alone apprimates the amino acid composition of the cyanobacteria.
- The amount of anthranilate and PABA precursors are equal to that of their products tryptophan and folate.
- Of the available precursors, 33% will go down our pathway. This is based off the idea that the enzymes may have similar affinities for the precursor.
- Folate is present in such small amounts, at 0.21 nanomoles of PABA, that its effect on acetaminophen production is neglegible [7].
These numbers show that there will probably be enough precursor to produce a useful, detectable quantity of acetaminophen. Based on literature and sequence estimates of aromatic amino acids, we can assume there would be at least that many moles of chorismate from which our added pathway pushes towards acetaminophen. The three calculations above can be averaged to finally predict 8.26mg ± 2.77mg acetaminophen per gram of Arthrospira platensis biomass.
We used different tryptophan estimates to reach several different predictions for acetaminophen, averaging 8.26mg ± 2.77mg acetaminophen per gram of Arthrospira platensis biomass or 8.26 µg per mL. This would be significantly above the limit of detection for our HPLC, at 50ng per ml, and serve as a starting point for optimizing production. This means that one 325mg dose of acetaminophen could be obtained in ~39g of biomass, meaning a 12 by 3 feet round pool could produce enough acetaminophen for more than 200 people every 10 days. While 39 grams isn't an ideal amount of medicine to consume, it does show that Arthrospira platensis has significant potential as a molecular factory for acetaminophen.
VITAMIN B12

The quantity of active form of B12 produced depends on a successful production and integration of the active B12 lower ligand, 5,6-dimethyl-benzimidazole (5,6-DMB). For phytoplankton in the wild, cobalt is often the limiting factor for growth and production of B12[11,12], while B12 production is limited by growth need in optimal media[12]. With ssuE and bluB genes inserted and regulated using a strong PrtC promoter, the activating lower ligand 5,6-DMB will be created in abundance[13]. Synechococcus and Arthrospira platensis both have CobS, bluB, and pGam genes that code for proteins which bind 5,6-DMB to the cobalt [14,15]. If these proteins work as well as in their origin organism, then assays report that 5,6-DMB has at least 100 times higher affinity for cobalt than the B12 analog ligand, adenine [16,17], meaning the DMB-B12 to B12 analog ration would be 100:1. Published HPLC results show that that Arthrospira platensis produces between 1.5-2.5µg B12 analogs per gram dry weight[18].
$$\frac{2.5µg\ B_{12}\ analog}{1g\ drymass} * \frac{100\ DMB\ bindings}{101\ DMB+adenine\ binding} = \frac{2.47µg\ active\ DMB-B_{12}}{1g\ biomass}$$An additional paper assayed Synechococcus elongatus sp WH 7803 at 10-18 moles per cell[16], and at a reported density 109 cells per liter (about a gram) [16,17], Synechococcus would produce 1.3 µg B12 per liter dry mass. Another older paper used microbiological assays along with TLC and HPLC, finding maximums of 2.4, 1.47, and 1.27 µg B12 per gram dry mass. Averaging the 6 data points and multiplying by a 100:1 conversion ratio results in a predicted production of 1.74 ±0.23µg active DMB-B12 per gram of Arthrospira platensis drymass, meaning the USDA’s recommended daily value of 6 µg could be obtained in one 3.5 gram serving.
Assumptions
- Gene inserts will be expressed, converting riboflavin-5′-phosphate to 5,6-DMB in excess [13].
- The bluB/CobS protein complex in Arthrospira platensis will attach 5,6-DMB to cobalt at rates similar to those assayed in Propionibacterium freudenreichii[14,15].
- Cobalt will be provided in excess of 0.3 mM according the BG-11 recipe, ensuring maximum precursor availability [16,17]
- Synechococcus 7942, 7803 and Arthrospira platensis will have similar rates of B12 production.
Future B12 projects might use chemo-trophic bacteria such as Methanosarcina barkeri which produces more than 1000 times more B12 and could be converted to active form using a similar bioengineering proccess[21]. One exceptional use of cyanobacterial B12 is growing cyanobacteria in the water used to grow rice, increasing the carbon fixation, nitrogen fixation, and bioenriching the rice with B12 [18].
BIOMASS

To understand the production capacity of our organism, we aggregated growth data from published papers and all of our lab’s growth data. Using carrying-capacity-limited logistic growth curves to fit our data to an equation, we modelled dried biomass and cell count with respect to time. We have also used growth optimization papers [9,10] to add additional dependent variables of temperature, light intensity, and starter culture density to our equation.
Light Intensity: μE m-2 s-1
Temperature: ℃
/* Starting Density: g biomass/ L */




