| Line 71: | Line 71: | ||
<p style="text-align:left !important;">After mini prep, we use the same restriction enzyme EcoR1 and Pst1 to do restriction digestion and gel electrophoresis. As seen, the size of K2197400 (2214 bp) and the backbone pSB1C3 (2070 bp)is almost the same, similar to our predicted result.</p> | <p style="text-align:left !important;">After mini prep, we use the same restriction enzyme EcoR1 and Pst1 to do restriction digestion and gel electrophoresis. As seen, the size of K2197400 (2214 bp) and the backbone pSB1C3 (2070 bp)is almost the same, similar to our predicted result.</p> | ||
<div class="row pop"> | <div class="row pop"> | ||
| + | |||
| + | |||
| + | |||
| + | <div class="row pop"> | ||
| + | <div class="col-xs-6"> | ||
| + | <a href="https://static.igem.org/mediawiki/2017/6/67/T--Hong_Kong_UCCKE--400abcdefghijklmnoph.jpg" style="width:100%;"/><img src="https://static.igem.org/mediawiki/2017/6/67/T--Hong_Kong_UCCKE--400abcdefghijklmnoph.jpg" style="width:100%;"/></a> | ||
| + | <br><span class="imgcaption">Project 400 (1)</span> | ||
| + | <br> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
<div class="col-xs-6"> | <div class="col-xs-6"> | ||
<a href="https://static.igem.org/mediawiki/2017/5/54/T--Hong_Kong_UCCKE--21asdfghj2.jpg" title="Image of gel electrophoresis of 400"><img src="https://static.igem.org/mediawiki/2017/5/54/T--Hong_Kong_UCCKE--21asdfghj2.jpg" style="width:100%;"/></a> | <a href="https://static.igem.org/mediawiki/2017/5/54/T--Hong_Kong_UCCKE--21asdfghj2.jpg" title="Image of gel electrophoresis of 400"><img src="https://static.igem.org/mediawiki/2017/5/54/T--Hong_Kong_UCCKE--21asdfghj2.jpg" style="width:100%;"/></a> | ||
| Line 88: | Line 100: | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 12:28, 31 October 2017
We have successfully cloned our plasmids into the E.coli. Then we did miniprep hence purified the plasmid. After that we did restriction map and have sent the parts to BGI for sequencing. The results are shown below.
Here are the results:
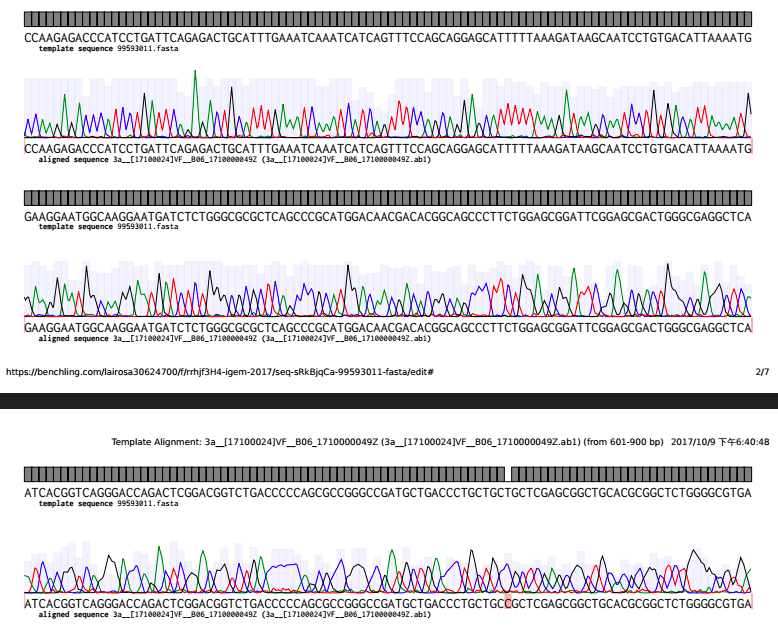
From the pictures below, the continuous grey line formed by rectangle boxes represents the theoretical sequence from the IDT,. Below it is the sequence of the gene we cloned. While there are flaws such as mutations along the gene we cloned (highlighted in red colour)
(Clicking on images will open the enlarged version in new tab)
Project 300
Restriction Map
After mini prep, we use the same restriction enzyme EcoR1 and Pst1 to do restriction digestion and gel electrophoresis. However, the size of K2197300 and the backbone pSB1C3 is almost the same, thus we cannot prove by the Gel photo.
Sequencing Result
With the help of HKCU, we sent our plasmid to BGI for sequencing. We aligned the sequence that we ordered from IDT(As the template sequence 99593011 below) with the sequencing result received from BGI(as the aligned sequence below). And here is our analysis.
The black area indicates that the two sequence are the same which those red strip represent that there're some mutations.
Project 400
Restriction Map
After mini prep, we use the same restriction enzyme EcoR1 and Pst1 to do restriction digestion and gel electrophoresis. As seen, the size of K2197400 (2214 bp) and the backbone pSB1C3 (2070 bp)is almost the same, similar to our predicted result.