| Line 14: | Line 14: | ||
To further verify our organism's amount of acetaminophen precursor, we ran both the genome and ribosomal protein sequences through a custom Python program converting codons to amino acids and calculating aromatic amino acid molar percentages which resulted in 9.3% and 5.14% respectively. | To further verify our organism's amount of acetaminophen precursor, we ran both the genome and ribosomal protein sequences through a custom Python program converting codons to amino acids and calculating aromatic amino acid molar percentages which resulted in 9.3% and 5.14% respectively. | ||
</br> | </br> | ||
| + | $$ \begin{align} | ||
| + | &\rightarrow LuxR \\ | ||
| + | LuxAHL+LuxR & \leftrightarrow RLux\\ | ||
| + | RLux+RLux &\leftrightarrow DRLux\\ | ||
| + | DRLux+P_{luxOFF} & \leftrightarrow P_{luxON}\\ | ||
| + | P_{luxON}&\rightarrow P_{luxON}+mRNA_{Bxb1}\\ | ||
| + | mRNA_{Bxb1}&\rightarrow Bxb1\\ | ||
| + | LuxAHL &\rightarrow \\ | ||
| + | LuxR &\rightarrow \\ | ||
| + | RLux &\rightarrow\\ | ||
| + | DRLux &\rightarrow\\ | ||
| + | mRNA_{Bxb1} &\rightarrow\\ | ||
| + | Bxb1 &\rightarrow | ||
| + | \end{align}$$ | ||
(Equation/ table summing amino acids converting to molar percentage) | (Equation/ table summing amino acids converting to molar percentage) | ||
</br> | </br> | ||
Revision as of 19:55, 20 September 2017
Modeling
Estimate and optimize yield.
Acetaminophen 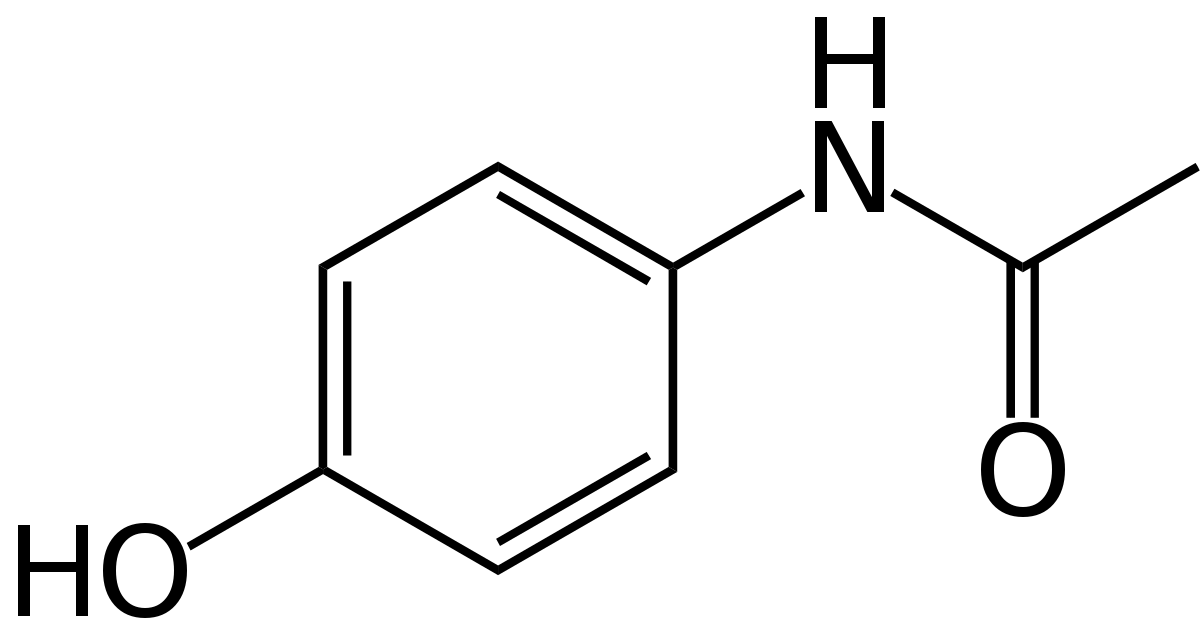
To predict theoretical acetaminophen production, we calculated the amount of a precursor, chorismate, by quantifying its main products, the aromatic amino acids phenylalanine, tyrosine, and tryptophan.
Since no amino acid composition data was available for Synechococcus, we started by using literature data for similar cyanobacteria species Spirulina and Synechocystis and found that between one and 13.6 percent of amino acids were aromatics by mass, or between XX and YY molar percent.
To further verify our organism's amount of acetaminophen precursor, we ran both the genome and ribosomal protein sequences through a custom Python program converting codons to amino acids and calculating aromatic amino acid molar percentages which resulted in 9.3% and 5.14% respectively.
$$ \begin{align}
&\rightarrow LuxR \\
LuxAHL+LuxR & \leftrightarrow RLux\\
RLux+RLux &\leftrightarrow DRLux\\
DRLux+P_{luxOFF} & \leftrightarrow P_{luxON}\\
P_{luxON}&\rightarrow P_{luxON}+mRNA_{Bxb1}\\
mRNA_{Bxb1}&\rightarrow Bxb1\\
LuxAHL &\rightarrow \\
LuxR &\rightarrow \\
RLux &\rightarrow\\
DRLux &\rightarrow\\
mRNA_{Bxb1} &\rightarrow\\
Bxb1 &\rightarrow
\end{align}$$
(Equation/ table summing amino acids converting to molar percentage)
Using our sequence generated median chorismate value of 9.3% and the assumption that our enzymes would take a third of the chorismate precursor, we got an estimate for acetaminophen concentration of between 17.5mg and 1.29 mg per gram dried biomass.
These numbers show that there is likely enough precursor and that acetaminophen production should be within a measurable range.
(Equation producing acetaminophen)
Biomass 
To understand the production capacity of our organism, we aggregated growth data from published papers and all of our lab’s growth data. Using limited logistic growth curves and linear algebra to fit our equation, we modelled dried biomass and cell count per time, with the additional dependent variables of temperature, light intensity, and starter culture density.
Timescale:
days
Light Intensity:
μE m-2 s-1
Temperature:
℃
Starting Density:
g biomass/ L
References
Growth optimization of Synechococcus elongatus PCC7942 in lab flasks and a 2-D photobioreactor Carbon metabolism and energy conversion of Synechococcus sp. PCC 7942 under mixotrophic conditions Characterization of the Role of para-Aminobenzoic Acid Biosynthesis in Folate Production by Lactococcus lactis 6S RNA: recent answers – future questions


