| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | <div class = "container"> | ||
| + | <h2 >The Problem</h2> | ||
| + | <ul> | ||
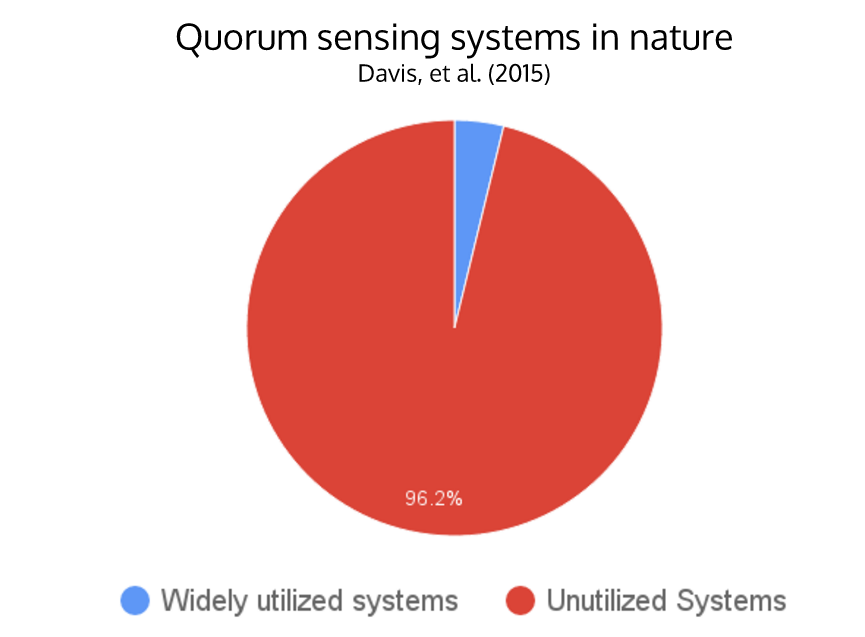
| + | <li>There are only around 4 widely-used N-acyl homoserine lactone (AHL) quorum sensing systems, while there are over 100 different systems found in nature</li> | ||
| + | <li>AHL molecules are not completely system-specific, as crosstalk can occur between systems-this causes problems with developing functional genetic circuits</li> | ||
| + | <li>AHL quorum sensing is used to regulate factors such as virulence and biofilm formation, potentially activating pathogenic bacteria</li> | ||
| + | </ul> | ||
| + | <div class = "center"> | ||
| + | <img src = "https://static.igem.org/mediawiki/2016/e/e8/T--Arizona_State--pie.png" alt = "piechart" style = "max-width: 600px; width: 100%"> | ||
| + | </div> | ||
| − | < | + | <h2>Brief Summary</h2> |
| − | + | ||
| − | + | <p> Quorum sensing is the ability of bacteria to scan their surroundings and detect concentrations of their own population. A species of bacteria will produce an inducer protein, which generates chemical signals in the form of N-acyl homo-serine lactones (AHLs). When the AHL reaches a high concentration, the bacterial cells will respond by collectively activating a set of genes. In nature, these AHLs are able to drastically influence the growth behavior of bacterial cells, activating biofilm formation, bioluminescence, virulence, motility, etc. </p> | |
| − | <p> | + | |
| + | <p> The objective of our project is to design and test a variety of quorum sensing networks. We have developed a flexible testing platform in which the QS system is separated into two components designated the “Sender” and the “Receiver”. The AHL synthase is expressed in the Sender cell, while the inducible promoter and regulator are carried by a Receiver cell. When the Sender produces a signal, the HSL, it diffuses across cell membranes and activates the Receiver. In our current system, Receivers will express green fluorescent protein (GFP) in response to induction by Senders from different bacterial species. Ideally, the designed systems would have low amounts of interference and form a functional genetic circuit. Our team has built 10 senders and their respective receivers. </p> | ||
| − | < | + | <p> Our iGEM team is investigating the diverse applications that fit with our quorum sensing project. Some of the sub-projects include: investigating the Aub strain, which originates from unidentified soil bacterium; writing a white paper on addressing potential safety concerns with AHLs, a visual optical density test on AHL production by Senders, a more comprehensive characterization of AHLs produced by our Senders using gas chromatography, a direct comparison induction test on Senders using the well-characterized part F2620, and develop comprehensive safety procedures for the handling of AHLs. The white paper and safe disposal protocol are several of our human practices projects that the ASU team investigated during the course of the project. </p> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | <h2>MOTIVATION</h2> |
| + | <p> N-acyl homoserine lactone (AHL) quorum sensing (QS) is a type of communication that allows bacteria to monitor their population density for the purpose of controlling various group activities such as virulence or luminescence. Synthetic biologists have taken advantage of the simplicity of the QS system to incorporate signal-processing pathways into genetic circuits. This project aims to determine pathways with minimal overlap (“crosstalk”) and engineer QS modules as a flexible tool for building layered genetic circuits. Crosstalk occurs when when a single regulator is activated by multiple varieties of HSL molecules, impeding successful operation of complex genetic circuitry that uses multiple quorum sensing pathways. Homologous AHL networks have been identified in over 100 species of bacteria, but only four have been used in synthetic systems reported to date. Our goal is to expand the QS toolbox and enable the implementation of higher-order, complex genetic circuitry in synthetic biology.</p> | ||
| − | < | + | <p>In addition to expanding the QS toolbox, our team felt that an in-depth investigation on the safety of AHLs would be valuable to future quorum sensing research. From the interviews and questionnaires that we conducted to industry experts, it was clear that AHLs were not a focal point in their safety reviews. However, from literature reviews, we found that there are many opportunistic pathogens that utilize AHL to activate virulence. A brief list is shown below:</p> |
| + | <center><img src="https://static.igem.org/mediawiki/2016/f/f4/T--Arizona_State--crosstalklist.png"></center> | ||
| + | <p>Because there are potential pathogens that could become activated, proper safety measures in the development and production of AHLs were necessary. In our Human Practices, we gathered information about the dangers of AHLs from experts and literature, developed a safe disposal plan, tested the safe disposal plan as an extension of our F2620 induction tests, and wrote a report on suggestions for the safety of AHLs. In addition, we added a safety section to our parts pages that we submitted to the registry. </p> | ||
| + | |||
| + | </div> | ||
| + | <br> | ||
| − | |||
| − | |||
| − | < | + | <div class="container"> |
| − | + | <h2>N-Acyl Homoserine Lactones</h2> | |
| − | </ | + | <p>AHL quorum sensing has a myriad of different systems. A total of 10 systems were investigated in this project </p> |
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | </p> | + | |
| + | <table style="width:100%"> | ||
| + | <tr> | ||
| + | <th>AHL System</th> | ||
| + | <th>Bacteria of Origin</th> | ||
| + | <th>AHL Name</th> | ||
| + | <th>3D-Model</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Aub</td> | ||
| + | <td>Unknown</td> | ||
| + | <td>N-(2-oxooxolan-3-yl)dodecanamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/3/38/T--Arizona_State--aubhsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Bja</td> | ||
| + | <td>Bradyrhizobium japonicum</td> | ||
| + | <td>3-methyl-N-[(3S)-2-oxooxolan-3-yl]butanamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/b/b0/T--Arizona_State--bjahsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Bra</td> | ||
| + | <td>Paraburkholderia kururiensis</td> | ||
| + | <td>(3S)-3-[(2-oxo-3-phenylpropyl)amino]oxolan-2-one</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/4/42/T--Arizona_State--brahsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Cer</td> | ||
| + | <td>Rhodobacter sphaeroides</td> | ||
| + | <td>(Z)-3-hydroxy-N-[(3S)-2-oxooxolan-3-yl]tetradec-7-enamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/6/6d/T--Arizona_State--cerhsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Esa</td> | ||
| + | <td>Erwinia stewartii</td> | ||
| + | <td>3-oxo-N-[(3S)-2-oxooxolan-3-yl]hexanamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/c/c5/T--Arizona_State--esahsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Las</td> | ||
| + | <td>Pseudomonas aeruginosa</td> | ||
| + | <td>3-oxo-N-(2-oxooxolan-3-yl)dodecanamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/e/ef/T--Arizona_State--lashsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Lux</td> | ||
| + | <td>Vibrio fischeri</td> | ||
| + | <td>3-oxo-N-(2-oxooxolan-3-yl)hexanamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/c/c5/T--Arizona_State--esahsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Rhl</td> | ||
| + | <td>Rhizobium leguminosarum</td> | ||
| + | <td>N-(2-oxooxolan-3-yl)butanamide</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/2/28/T--Arizona_State--rhlhsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Rpa</td> | ||
| + | <td>Rhodopseudomonas palustris</td> | ||
| + | <td>(S)-α-amino-γ-butyrolactone</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/0/08/T--Arizona_State--rpahsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Sin</td> | ||
| + | <td>Sinorhizobium meliloti</td> | ||
| + | <td>N-[(3S)-2-oxooxolan-3-yl]octanamide*</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2016/6/6d/T--Arizona_State--sin2hsl3d.png" height=150px></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p>*Sin system produces 6 different variants of AHL. The 3D structures of all the Sin compounds can be found <a href="https://www.dropbox.com/sh/cfo744xq81t6iu4/AAAAG8yiFkHfnUojBZE7bWKla?dl=0">here</a>.</p> | ||
| + | <p>AHLs share the same basic structure, with a lactone ring, an N-acyl and ketone group. The defining R group lies in the acyl tail, which is the primary determinant in its transcription factor binding affinity. The graphic below demonstrates the categorization of the AHLs produced by the 10 studied systems</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2016/4/4f/T--Arizona_State--ahlcategory.png" height=500px></center> | ||
| + | <h2>F2620 Inductions</h2> | ||
| + | <p>The ASU team would like this experiment to be considered for the Gold Medal Requirement of improving the characterization of a previously existing BioBrick part. All 10 systems were studied in an induction test. The part BBa_F2620 (designed by Barry Canton from MIT) was used to induce production in the Lux AHL system and test induction in any other AHL systems. By studying interactions between the 10 constructed Senders and F2620, we were able to analyze the systems for potential orthogonality. The resulting part collection allows direct comparison in AHL induction between multiple systems. The Part Numbers for these Senders are | ||
| + | <a href="http://parts.igem.org/Part:BBa_K2033000">BBa_K2033000</a>, | ||
| + | <a href="http://parts.igem.org/Part:BBa_K2033002">BBa_K2033002</a>, | ||
| + | <a href="http://parts.igem.org/Part:BBa_K2033004">BBa_K2033004</a>, | ||
| + | <a href="http://parts.igem.org/Part:BBa_K2033006">BBa_K2033006</a>, | ||
| + | <a href="http://parts.igem.org/Part:BBa_K2033008">BBa_K2033008</a>, which correspond to the Aub, Bja, Bra, Cer, and Sin systems. We believe that this Part Collection is a valuable addition to the registry, because while small in size, it contributes 5 Senders to the registry which contains around 6 Senders. In addition, this will add information about these new systems to a well-characterized and widely-used part in F2620. Not only will this determine interactions between systems, but also, provide characterization information for any future quorum sensing research involving those systems.</p> | ||
| + | <p>We also created Senders for the Esa, Las, Lux, Rhl, and Rpa systems, which already exist in the registry. These correspond to the following parts:</p> | ||
| + | <ul> | ||
| + | <li>EsaI-<a href="http://parts.igem.org/Part:BBa_K1670004">BBa_K1670004</a></li> | ||
| + | <li>LasI-<a href="http://parts.igem.org/Part:BBa_C0178">Bba_C0178</a></li> | ||
| + | <li>LuxI-<a href="http://parts.igem.org/Part:BBa_C0161">BBa_C0161</a></li> | ||
| + | <li>RhlI-<a href="http://parts.igem.org/Part:BBa_C0170">BBa_C0170</a></li> | ||
| + | <li>RpaI-<a href="http://parts.igem.org/Part:BBa_K1421006">BBa_K1421006</a></li> | ||
| + | <p>We added the induction data that we collected for these systems to their respective parts pages, which provides information on their compatibility with F2620.</p> | ||
</div> | </div> | ||
| + | <div class = "container"> | ||
| + | <br> | ||
| − | < | + | <br> |
| + | <h2>REFERENCES</h2> | ||
| + | <ol> | ||
| + | <li>Davis, René Michele, Ryan Yue Muller, and Karmella Ann Haynes. "Can the Natural Diversity of Quorum-Sensing Advance Synthetic Biology?" Frontiers in Bioengineering and Biotechnology 30th ser. 3 (2015)</li> | ||
| + | <li>Borchardt, S.A., Allain, E.J., Michels, J.J. “Reaction of Acylated Homoserine Lactone Bacterial Signaling Molecules with Oxidized Halogen Antimicrobials” Applied and Environmental Microbiology. 67.7. (2001); 3174-3179. </li> | ||
| + | <li>Canton, Barry, Anna Labno, and Drew Endy. "Refinement and Standardization of Synthetic Biological Parts and Devices." Nature Biotechnology 26.7 (2008): 787-93.</li> | ||
| + | <li>Scott, Spencer R., and Jeff Hasty. "Quorum Sensing Communication Modules for Microbial Consortia." ACS Synthetic Biology 5.9 (2016): 969-77.</li> | ||
| + | <li>Miller, Melissa B., and Bassler, Bonnie L. “Quorum Sensing in Bacteria.” Microbiology. 55. (2001); 165-99</li> | ||
| + | <li>Tait, Karen, Zoe Hutchison, Fabiano L. Thompson, and Colin B. Munn. "Quorum Sensing Signal Production and Inhibition by Coral-associated Vibrios." Environmental Microbiology Reports 2.1 (2010): 145-50.</li> | ||
| + | <li>Grant, Paul, Dalchau, Neil, Brown, James R. “Orthogonal intercellular signaling for programmed spatial behavior.” Molecular Systems Biology 12.1 (2016):849. </li> | ||
| + | <li>Steindler, Laura, Bertani, Iris, De Sordi, Luisa. "LasI/R and RhlI/R Quorum Sensing in a Strain of Pseudomonas aeruginosa Beneficial to Plants." Applied and Environmental Microbiology. 75.15: 5131–5140. </li> | ||
| + | <li>Lindemann A. "Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum." PubMed. 108.40 (2011):16765-70.</li> | ||
| + | <li>Ahlgren NA. "Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia." PubMed. 108.17 (2011):7183-8..</li> | ||
| + | <li>Hoang, Hanh H. "The LuxR Homolog ExpR, in Combination with the Sin Quorum Sensing System, Plays a Central Role in Sinorhizobium meliloti Gene Expression." Journal of Bacteriology. 186.16 (2004): 5460-5472. </li> | ||
| + | <li>Marketon, Melanie M. "Characterization of the Sinorhizobium meliloti sinR/sinI Locus and the Production of Novel N-Acyl Homoserine Lactones." Journal of Bacteriology. 184.20 (2002): 5686-5695.</li> | ||
| + | <li>Case, Rebecca J., Labbate, Maurizio, Kjelleberg, Staffan. "AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria." The ISME Journal. 2 (2008): 345–349.</li> | ||
| + | <li>Winstanley, Craig, Fothergill, Joanne L. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections." FEMS Microbiology Letters. (2009) 1-9. </li> | ||
| + | <li>Watson, William T. "Structural Basis and Specificity of Acyl-Homoserine Lactone Signal Production in Bacterial Quorum Sensing." Molecular Cell. 9.1 (2002) 685–694.</li> | ||
| + | </ol> | ||
| − | |||
| − | |||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
</html> | </html> | ||
| + | {{Arizona_State_Footer}} | ||
Revision as of 18:55, 1 August 2017