EXPERIMENTS:
STEP 1: Create guideRNA Plasmid
Why
We produced our gRNA plasmids in order to repress targeted fluorescent proteins on our reporter plasmid. This allows more combinations than possible with reporters alone.
How
Refer to step 2 for plasmid construction. The sgRNA plasmid uses static promoter J23119, and sgRNA in place of reporter proteins. We constructed plasmids that contain one gRNA each, although we could expand this to include 3 or more gRNAs per plasmid. In our lab book you can see that we can easily transform this plasmid into E. coli along with pReporter with high efficiency. The gRNA plasmid uses the same ligation strategy as pReporter which means that the process of creating more combinations is simple and minimises the cost associated with many enzymes.
Results
Refer to CRISPRi & gRNA efficiency results.
STEP 2: Create Reporter Plasmid

Why
To create a stepwise and logical order of production, a reporter plasmid was constructed firstly, out of only various promoter types, mRFP and terminators, to allow for testing. This was then escalated to include dCas9 for the final experiment.
How

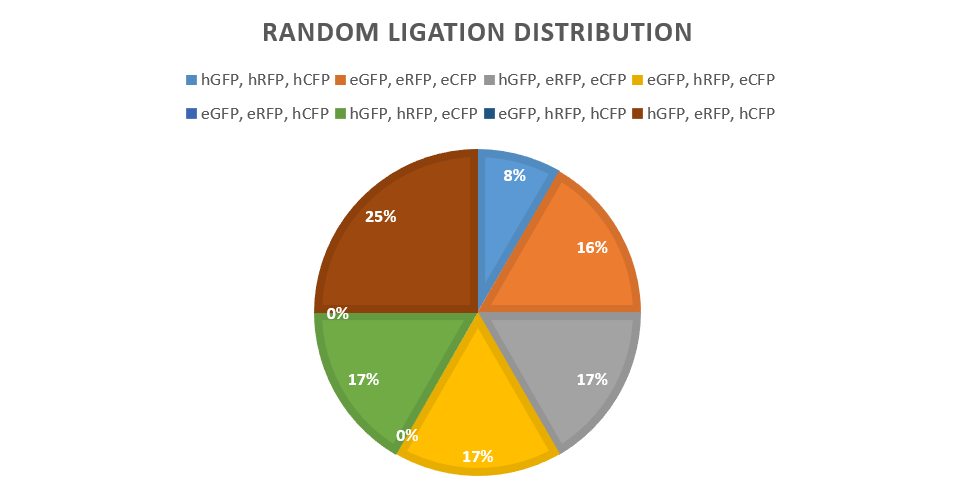
Each individual Promoter-Reporter-Terminator brick contains interchangeable parts. The three parts are linked together with Bsa1 sites so that there is no preference for any part when ligating together. This allows randomness to be added later. This method is used also for the construction of Promoter-gRNA-Terminator bricks so that this could be randomised in the future. The bricks are then flanked by a prefix and suffix, and these are flanked by restriction sites ABCD on either end. Digestion of bricks with A+B, B+C, and C+D allows any brick to be placed in any position within the plasmid but it would be pre-determined. This means that the no one promoter-reporter-terminator brick would be limited to one specific place in the plasmid, which allows another level of randomness in assembly as we would not know which reporter was being placed where, which could also affect expression levels.
The arrangement of the DCBA restriction system means that any brick can be placed in any position in the plasmid which allows expansion of possibilities whilst maintaining randomness of insertion later. Bricks can be joined together via amplifying each randomly assembled brick through common amplification sites and then cutting them using a set of restriction enzymes which give each plasmid a specific order of bricks, depending on which are cut and then ligated together.

This was then used for the promoter library.
After this, the same method as above was used for the final dCas9 including vector. However with an addition of dCas9 in the final ligation phase. In this way two promoter libraries were produced.
Results
Found in promoter library section.
STEP 3: Promoter Library

Why
To establish the relative strength of every single promoter, each of them must be tested and their expression level measured throughout the bacterial growth. This will provide data to demonstrate it is possible to differentiate between them, proving that our key will work effectively by having identifiable spectral states. Getting unique spectral profiles is a cause of concern as the excitation and emission spectra of RFP, CFP and GFP overlap somewhat and cross excitation will occur, perhaps masking CFP’s emissions and reducing the potential for separate keys to be identifiable. To fully address this, a promoter library was constructed to assess the promoter’s strength.
How
For the full description, refer to the lab book in week. To summarise, overnight cultures of each specimen were inoculated into a new batch of media. These were then grown at 37°C for 24 hours, with measurements of OD600nm absorbance and fluorescence carried at 0, 2, 4, 6, and 24 hours. The promoters were paired with a downstream RFP reporter gene.
Results
| Figure 1. This graph shows the relative fluorescent intesnity of strong and weak RBS promoters SP1-E and WP1-4. These were grown for a 24 hour period and measurements were taken at 0, 2, 4, 6, 9, and 24 hours. |
|---|