|
|
| Line 1: |
Line 1: |
| − | {{Aix-Marseille|title=Engineered M13 modelling|toc=__TOC__}} | + | {{Aix-Marseille|title=Modelling PLP production|toc=__TOC__}} |
| | | | |
| − | <!-- commentaire 1: pour le wiki pas besoin de htlm -->
| + | [[File:T--Aix-Marseille--ModelM13.png|400px|right|thumb|To produce PLPs, bacteria must contain both a helper phage (like M13KO7), that codes for the different phage proteins, and also a phagemid, that encodes the toxic gene we want to include in the PLP. |
| − | <!-- Titre entre = (plus tu en met moins le titre est important -->
| + | If M13KO7 is packaged a replicative phage is produced, which we do not want, and if the phagemid it packaged then a PLP is produced.]] |
| − | <!-- GRAS entre ''' Italique entre '' -->
| + | |
| − | <!-- intra lien [[Page (debut par team)|nom]] -->
| + | |
| − | <!-- pour les images. Pour les mettre sur le sites : wiki tools/Upload files. Après tu sélectionnne ton image et tu la nomme par T--Aix-Marseille--NOM.png/.jpg-->
| + | |
| − | <!-- Pour les mettre sur la pages : [[File:T--Aix-Marseille--NOM.png|TAILLE(XXXpx)|right/Left/center|thumb|Legende]]-->
| + | |
| | | | |
| − | [[Team:Aix-Marseille/Model2|James Proposal]]
| + | '''The problem''' |
| | | | |
| − | [[File:T--Aix-Marseille--ModelM13.png|400px|right|thumb|In order to produce PLPs, both M13KO7 (produces all the phage proteins) and the phagemid (containing the toxin gene) need to be integrated into ''E.coli''. During the process, if p5 packages M13KO7 it will produce a replicative phage and if it packages our phagemid it will produce PLPs.]] | + | Our objective is to produce [[Team:Aix-Marseille/Bacteriophages|phage like particles (PLP)]], |
| | + | for this, the bacteria must contain both a helper phage and also a phagemid (see figure). |
| | + | During phage production, several key events determine which DNA molecules are packaged into phage or PLP. |
| | + | These involve recognition of the [[Team: Aix-Marseille/M13|M13]] replication origin by several phage proteins. |
| | | | |
| − | == Introduction ==
| + | A major hurdle to marketing '''KILL XYL''' is obtaining the necessary authorizations. |
| | + | Our [[Team:Aix-Marseille/HP/Interviews|interviews]] and [[Team:Aix-Marseille/Legislation|legislation]] study |
| | + | both showed that the number of viable phages could be a problem. |
| | + | We therefore decided to [[Team:Aix-Marseille/M13_test|measure]] and model the ratio between viable phage and phage-like particles, and so try to optimize this ratio to facilitate the preparation of pure PLP. |
| | | | |
| − | Modelling is an essential aspect of engineering, and we wanted to engineer a novel specific phage targeted to [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']]. Apart from a preliminary exercise model used to test our understanding of our system and train ourselves in modelling, most of our work was done using a (heavily) adapted version of an already published model of [[Team:Aix-Marseille/M13|M13]] physiology.
| + | '''The Model''' |
| | | | |
| − | Modelling allows engineers to explore several avenues of research very quickly and to discard those less likely to succeed early on and at all stages of prototyping and development. For this back and forth betwen wet- and dry-lab to be meaningfull, it needs to be validated against preliminary data, which we did by ensuring our model fitted experimental particle profiles obtained by the wetlab team. We then investigated wether levels of certain key proteins changed particle composition, before determining the ideal number of plasmid copies to optimize our particle production.
| + | We based our model on a recently published model of "wild-type" M13 replication |
| | + | <ref name=Smeal>Smeal ''et al'', Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017</ref> |
| | + | <ref>Smeal ''et al'', Simulation of the M13 life cycle II: Investigation of the control mechanisms of M13 infection and establishment of the carrier state, Virology, 500, January 2017</ref>. |
| | + | <!-- Erwan check this and fix it --> |
| | + | This model was modified to incorporate: |
| | + | The helper-phage E. coli plasmid origin; |
| | + | the modification of the helper-phage M13 origin; |
| | + | the presence of a phagemid, with its own replicative origin and M13 origin. |
| | + | These modifications added xxx species, zzz equations and yyy parameters to the original model, and they modified www parameters of the original model. |
| | + | The modified model is available [[File:T--Aix-Marseille--Model.zip|here.]] <!--it needs to be uploaded onto the iGEM server-->. |
| | | | |
| − | Our project, [[Team:Aix-Marseille/Project|'''KILL XYL''']], sets to engineer a cure against the disease caused by the plant pathogen [[Team:Aix-Marseille/Xylella_fastidiosa|''Xylella fastidiosa'']]. One of our approaches is to engineer [[Team:Aix-Marseille/Bacteriophages|phage-like particles]] (PLPs), that will deliver toxic genes into the bacterium. Phage systems, like [[Team:Aix-Marseille/M13|M13]], have been employed in biotechnological applications, most prominently in the identification and maturation of medically-relevant binding molecules through phage display <ref>Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat Rev Micro 13, 777–786 (2015).</ref>.
| + | We used the initial [[Team:Aix-Marseille/M13_test|measurement]] to constrain parameters of the model, |
| | + | along with published numbers for the number of copies of plasmids with different replicative origins. |
| | + | <!--What assumptions remain in the model--> |
| | | | |
| − | [[Team:Aix-Marseille/M13|M13]] has a particular life-cycle among phages: it is neither a virulent nor a temperate phage but rather engages in chronic infection. One of the crucial events of it's multiplication is the packaging of its own DNA in the capsid. To do so, [[Team:Aix-Marseille/M13|M13]]'s ''protein 5'' (p5) targets M13 origin of replication (M13ori) and packages the single-strand DNA. This step is crucial to initiate formation of the phage particle. Afterwards, an assembly complex (made of proteins p1, p4 and p11) assembles the capsid (made of proteins p3, p6, p7, p8, p9) around the ssDNA. P3 and p6 are of particular importance, since they close the phage's capsid which provokes it's liberation in the environment.
| + | '''Results''' |
| | | | |
| − | One of the major reasons we chose to use [[Team:Aix-Marseille/Bacteriophages|phage-like particles (PLPs)]] was for our device to be non replicative, to prevent the spread of GMOs in the environnement. We [[Team:Aix-Marseille/M13_Design|designed]] these PLPs by using a modified M13's genome to prevent replication and by adding a phagemid in the factory ''E.coli'': our factory bears ''two plasmids'', a modified M13 genome (M13KO7) with several genes inserts in it M13ori and a phagemid with a M13ori.
| + | We need one or two graphs here showing how the packaging ratio depends on three main parameters: the DNA III polymerase binding rate to the E-coli origin of replication (Figure 2) of either the plasmid or phagemid, the initial ratio of transfected phagemid and helper phage (Figure 3), and the difference in p5 affinity for eitheir plasmid's M13 ori (Figure 4). |
| | + | We also observed increasing the number of transfected plasmids increased production, up to a certain point, which we determined (Figure 5). |
| | | | |
| − | ==Our model==
| + | A description of these graphs. |
| | | | |
| − | <!--les réference se place entre <ref>xxx</ref>. Si tu souhaite mettre plusieur fois la meme ref il faut que tu mettes <ref name=nom>xxx</ref>. Sinon il va croire que c'est des ref differentes-->
| + | '''Conclusions''' |
| | | | |
| − | After learning the ropes using an overly simplified model (which didn't amount to much), we decided to adopt the model of a "wild-type" M13's biology <ref name=Smeal>Smeal ''et al'', Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017</ref><ref>Smeal ''et al'', Simulation of the M13 life cycle II: Investigation of the control mechanisms of M13 infection and establishment of the carrier state, Virology, 500, January 2017</ref> to fit our own engineered version of M13 and better inform wet lab experiments, as well as explain some of our preliminary findings.
| + | <!--What we write here determines if you get a gold medal or not--> |
| − | | + | Clearly the initial design needs to be improved to increase the proportion of PLP produced. |
| − | Smeal's ''et al'' model describes the whole physiology of M13 in an infected cell. We conserved the parts of their model which deals with cellular processes such as DNA replication and transcription as well as protein translation, as there is no reason to think our design would affect these processes in a significant manner since those are physiological cellular processes.
| + | The model has shown as that the parameters of prime importance for determining the proportion of PLP produced are .... <!--This corresponds to the figures 2 through 5. |
| − | | + | So to implement our project it will be necessary to redesign certain features. Specifically <!-- What goes here -->. |
| − | However, we heavily altered the parts describing the interactions between the phage's proteins and genetic elements, since these are the diverging points between our construct and the wild-type M13.
| + | |
| − | We accounted for the presence of two plasmids: M13KO7 and the lethal phagemid to be packaged, which fits our [[Team:Aix-Marseille/M13_Design|design]] by incorporating all the related pathways. We adapted the parameters to account for the differences between our constructs and wild-type M13.
| + | |
| − | We also accounted for the possibility, in case of insufficient p3 or p6 concentrations, for the capsid to incorporate another plasmid. This translated by adding another step between capsid formation and phage release.
| + | |
| − | | + | |
| − | ===Model validation===
| + | |
| − | We used the model we devised to predict a particule profile (i.e. ratio of replicative phages vs. PLPs) compared it to the particle profile obtained in wetlab:
| + | |
| − | | + | |
| − | This can perhaps be explained by two phenomena:
| + | |
| − | | + | |
| − | # Our test method would label phages containing both M13KO7 and the phagemid as replicative phages. Perhaps several plasmids are incorporated in each particle, which would explain these results. See "Insufficient p3 and p6" for further exploration of this topic.
| + | |
| − | # Even when deleting the extra step allowing the capsid to incorporate several plasmids, we obtain 2/3 replicative particles and 1/3 PLPs. See "Ideal number of plasmid copies" for further exploration of the phenomena.
| + | |
| − | | + | |
| − | ==Insufficient p3 and p6==
| + | |
| − | P3 and p6 are proteins on the end of the capsid, which they "close", thereby provoking the liberation of the phage (or PLP) from the cell membrane. In the event of low p3 and p6 concentration, one can imagine capsid formation to continue with a second plasmid.
| + | |
| − | Our model takes this phenomena into account, which allows him to fit the experimental data with much more precision than without. This suggests it might be part of the cause for our less-than-optimal production of PLPs.
| + | |
| − | | + | |
| − | We would therefore suggest to engineer stronger promoters for p3 and p6 genes, to maximize their cytoplasmic concentration and diminish this unwanted phenomena.
| + | |
| − | | + | |
| − | ==Ideal number of plasmid copies==
| + | |
| − | Our Human Practice research makes it very clear our device cannot be replicative if it is to be used outside of the lab. We want to know the ideal number of plasmid copies to optimize production of PLPs over phages.
| + | |
| − | | + | |
| − | Our results show that, counter-intuitively, a higher-copy plasmid is actually '''less''' likely to be incorporated into a particle. We hypothesize this is because a higher-copy plasmid's ORI recruits DNA polymerase with more efficiency, which diminishes available ssDNA for p5 to bind to and form particles with.
| + | |
| − | | + | |
| − | We would therefore suggest to engineer a phagemid based on a lower-copy backbone, such as pSC101 or pBlueScript, to optimize production of PLPs. Following this directive, we tried to create a PLP based on pBlueScript which was more efficient than with pSB1C3.
| + | |
| − | | + | |
| − | ==Optimized system==
| + | |
| − | We ran our model with parameters reflecting the changes we propose and obtained the following particle profile:
| + | |
| − | | + | |
| − | ==Materials and Methods==
| + | |
| − | <!--Je pense approprié de créer une nouvelle page pour ça et de mettre cette partie dedans--> | + | |
| − | | + | |
| − | Our modelling was done using Matlab's Simbiology. The full .sbproj file (Simbiology file) can be found here. <!--ajouter fichier-->
| + | |
| − | | + | |
| − | ===Major parameters adaptation===
| + | |
| − | '''Polymerase binding event''': 2 times as likely to bind to phagemid (pUC ori) as opposed to M13KO7 (p15A ori). 1/10th times as likely to bind to pSC101 ori as opposed to M13KO7. These parameters were chosen because they conducted to copy numbers best fitting known ratios in the absence of incorporation into phage particles.<ref>http://blog.addgene.org/plasmid-101-origin-of-replication</ref>
| + | |
| − | | + | |
| − | | + | |
| − | ==References==
| + | |
Modelling PLP production
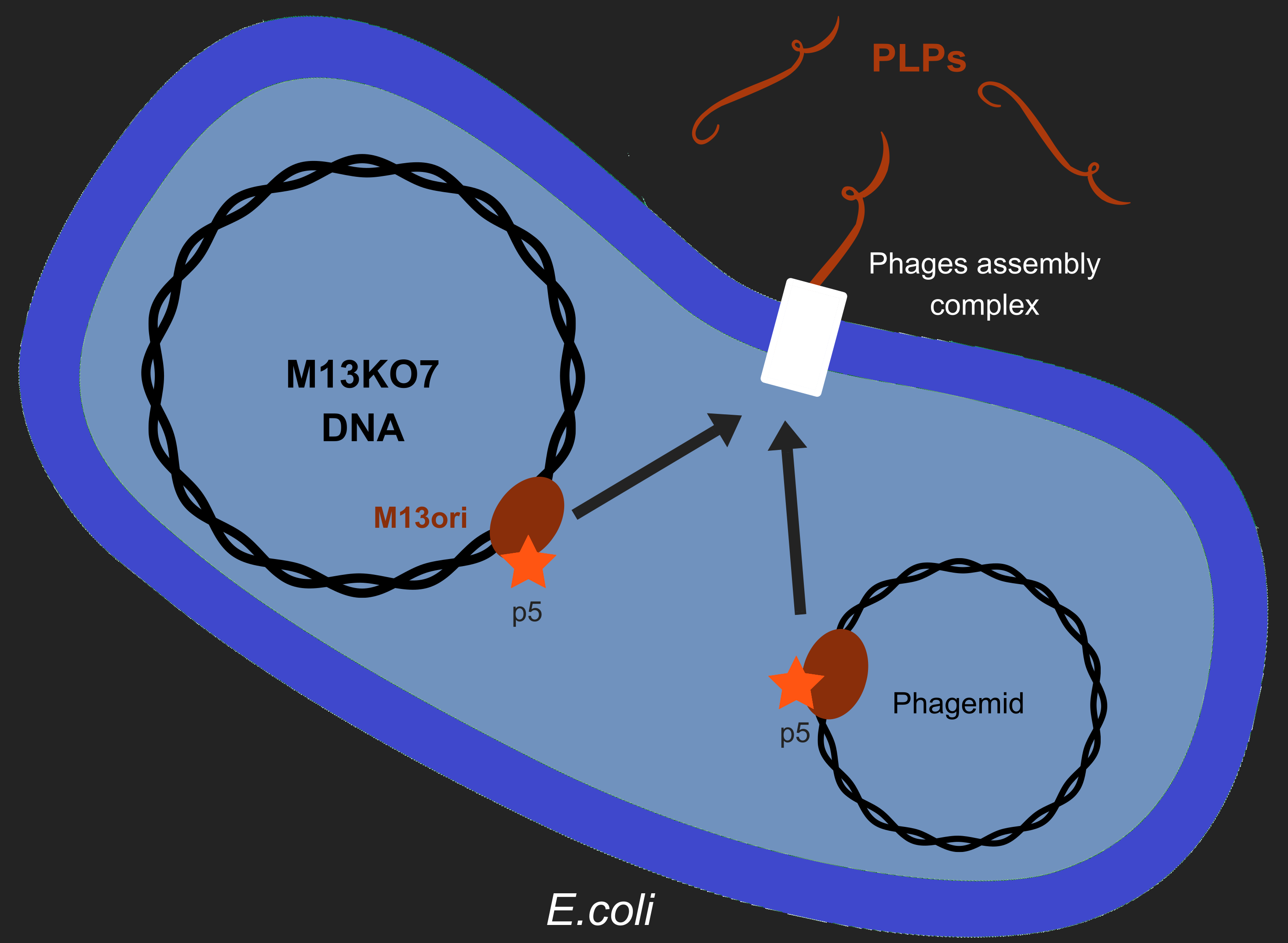
To produce PLPs, bacteria must contain both a helper phage (like M13KO7), that codes for the different phage proteins, and also a phagemid, that encodes the toxic gene we want to include in the PLP. If M13KO7 is packaged a replicative phage is produced, which we do not want, and if the phagemid it packaged then a PLP is produced.
The problem
Our objective is to produce phage like particles (PLP),
for this, the bacteria must contain both a helper phage and also a phagemid (see figure).
During phage production, several key events determine which DNA molecules are packaged into phage or PLP.
These involve recognition of the M13 replication origin by several phage proteins.
A major hurdle to marketing KILL XYL is obtaining the necessary authorizations.
Our interviews and legislation study
both showed that the number of viable phages could be a problem.
We therefore decided to measure and model the ratio between viable phage and phage-like particles, and so try to optimize this ratio to facilitate the preparation of pure PLP.
The Model
We based our model on a recently published model of "wild-type" M13 replication
[1]
[2].
This model was modified to incorporate:
The helper-phage E. coli plasmid origin;
the modification of the helper-phage M13 origin;
the presence of a phagemid, with its own replicative origin and M13 origin.
These modifications added xxx species, zzz equations and yyy parameters to the original model, and they modified www parameters of the original model.
The modified model is available File:T--Aix-Marseille--Model.zip .
We used the initial measurement to constrain parameters of the model,
along with published numbers for the number of copies of plasmids with different replicative origins.
Results
We need one or two graphs here showing how the packaging ratio depends on three main parameters: the DNA III polymerase binding rate to the E-coli origin of replication (Figure 2) of either the plasmid or phagemid, the initial ratio of transfected phagemid and helper phage (Figure 3), and the difference in p5 affinity for eitheir plasmid's M13 ori (Figure 4).
We also observed increasing the number of transfected plasmids increased production, up to a certain point, which we determined (Figure 5).
A description of these graphs.
Conclusions
Clearly the initial design needs to be improved to increase the proportion of PLP produced.
The model has shown as that the parameters of prime importance for determining the proportion of PLP produced are .... .
- ↑ Smeal et al, Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation, Virology, 500, January 2017
- ↑ Smeal et al, Simulation of the M13 life cycle II: Investigation of the control mechanisms of M13 infection and establishment of the carrier state, Virology, 500, January 2017


