Joe McKeown (Talk | contribs) |
Joe McKeown (Talk | contribs) |
||
| Line 34: | Line 34: | ||
<h2>In the Beginning...</h2> | <h2>In the Beginning...</h2> | ||
<p style="font-size: 18px;">When the York iGEM 2017 team first got together, we knew that co-culturing of microorganisms is becoming an extremely promising approach. In particular, this is true with regards to understanding natural and synthetic cell population interactions and applications in Industrial Biotechnology (e.g. manufacturing and drug research). We planned to use a co-culture comprising <em>Chlamydomonas reinhardtii</em> and <em>Escherichia coli</em> in order to create a somewhat self-sustaining bioreactor that could cost-efficiently produce biofuels. In such an instance, <em>C. reinhardtii</em> would be engineered to export maltose, which <em>E. coli</em> could use in its production of biofuel. Since the crux of this challenge lay in ensuring that <em>C. reinhardtii</em> would export maltose, we planned to use a strain of <em>E. coli</em> that produces ethanol, rather than over-complicate the experiment.</p> | <p style="font-size: 18px;">When the York iGEM 2017 team first got together, we knew that co-culturing of microorganisms is becoming an extremely promising approach. In particular, this is true with regards to understanding natural and synthetic cell population interactions and applications in Industrial Biotechnology (e.g. manufacturing and drug research). We planned to use a co-culture comprising <em>Chlamydomonas reinhardtii</em> and <em>Escherichia coli</em> in order to create a somewhat self-sustaining bioreactor that could cost-efficiently produce biofuels. In such an instance, <em>C. reinhardtii</em> would be engineered to export maltose, which <em>E. coli</em> could use in its production of biofuel. Since the crux of this challenge lay in ensuring that <em>C. reinhardtii</em> would export maltose, we planned to use a strain of <em>E. coli</em> that produces ethanol, rather than over-complicate the experiment.</p> | ||
| − | <p style="font-size: 18px;">We soon became acutely aware that, along with the maintenance of stable and productive co-cultures being technically challenging | + | <p style="font-size: 18px;">We soon became acutely aware that, along with the maintenance of stable and productive co-cultures being technically challenging, the usual methods of counting cells are often inaccurate, expensive, slow or destructive when applied to cultures of more than one organism. Thence, we have come to consider the desperate need for a Quicker Way to Analyse Co-Cultures (QWACC) a problem that must be rectified in order to advance the possibilities of research with co-cultures. We have not, however, abandoned the idea of producing cheaper biofuels with co-cultures and the <em>C. reinhardtii/E. coli</em> co-culture remains a centre piece of this project. This serves as a stark and ever-present reminder that the hardware and software we have developed should be easily used in conjunction with actual experiments involving co-cultures.</p> |
<br style="line-height: 10px;"> | <br style="line-height: 10px;"> | ||
</div> | </div> | ||
Revision as of 20:56, 6 September 2017
Upon being asked to find the number of microorganisms in a given sample, the response from members of our iGEM team has invariably been a long sigh and a plea to the realms of the supernatural.
That most human quality - laziness - alongside flaws in traditional techniques, which we will revisit below, has led to us designing a potential solution in the form of a Digital Inline Holographic Microscope (DIHM).
In particular, we discovered that, when studying co-cultures, there are precious few methods of counting cells that are accurate, fast, cheap and neither invasive, nor destructive.
We decided, therefore, to try to bring together two promising areas of science in this project:
Digital Holographic Microscopy and Co-Culturing.
In the Beginning...
When the York iGEM 2017 team first got together, we knew that co-culturing of microorganisms is becoming an extremely promising approach. In particular, this is true with regards to understanding natural and synthetic cell population interactions and applications in Industrial Biotechnology (e.g. manufacturing and drug research). We planned to use a co-culture comprising Chlamydomonas reinhardtii and Escherichia coli in order to create a somewhat self-sustaining bioreactor that could cost-efficiently produce biofuels. In such an instance, C. reinhardtii would be engineered to export maltose, which E. coli could use in its production of biofuel. Since the crux of this challenge lay in ensuring that C. reinhardtii would export maltose, we planned to use a strain of E. coli that produces ethanol, rather than over-complicate the experiment.
We soon became acutely aware that, along with the maintenance of stable and productive co-cultures being technically challenging, the usual methods of counting cells are often inaccurate, expensive, slow or destructive when applied to cultures of more than one organism. Thence, we have come to consider the desperate need for a Quicker Way to Analyse Co-Cultures (QWACC) a problem that must be rectified in order to advance the possibilities of research with co-cultures. We have not, however, abandoned the idea of producing cheaper biofuels with co-cultures and the C. reinhardtii/E. coli co-culture remains a centre piece of this project. This serves as a stark and ever-present reminder that the hardware and software we have developed should be easily used in conjunction with actual experiments involving co-cultures.
Old Science, New Tricks: DIHM
The first half of our bipartite project is the development of an inexpensive DIHM and accompanying software which is able to automatically count cells. Due to the inherently digital nature of this type of microscopy, software can easily be created and adapted such that the DIHM can not only count organisms but, also, differentiate between those that are distinguishable by physical appearance. Among the most important motivators of this hardware/software combination were speed of measurement (our target was the order of seconds or minute per measurement) and low costs for setup and maintenance. Further, in co-cultures wherein neither organism contains a fluorescent marker, the process of counting each type can become a rather complex endeavour. With our Quicker Way to Analyse Co-Cultures (QWACC), however, there exists the potential for real-time counting of all physically distinct organisms within a sample.
In the table, below, various methods of cell counting are compared with each other. This is not quantitative and simply shows how each technique stacks up compared to the rest of those on the list. This is denoted through a traffic light system - green indicates that the technique is desirable for the given quality, yellow corresponds to a reasonably desirable quality and red indicates that the technique is undesirable with respect to the quality.

- Table 1: A qualitative comparison of organism counting techniques. Green: desirable; yellow: reasonable; red: undesirable.
Old Science, New Tricks: Co-Culture
The second half of the project involves the creation of a co-culture comprising Escherichia coli and Chlamydomonas reinhardtii. Since E. coli and C. reinhardtii are separated in size by an order of magnitude, it is possible to use so-called blob detection algorithms to locate, count and distinguish between the two organisms in holograms formed by a DIHM.
Digital Inline Holographic Microscopy
How does it work?
Digital holographic microscopy makes use of diffraction. This a physical phenomenon wherein objects (or apertures) in the path of a source of waves will create patterns in the waves. These patterns depend on the shape of the objects themselves. Since electromagnetic waves are, somewhat unsurprisingly, waves, it is possible to use the diffraction of light to create patterns that we can see.
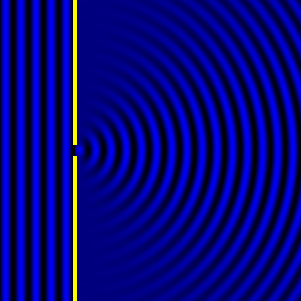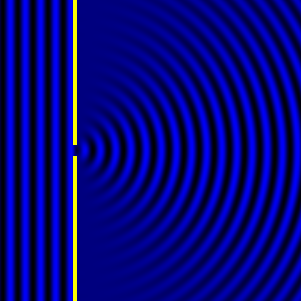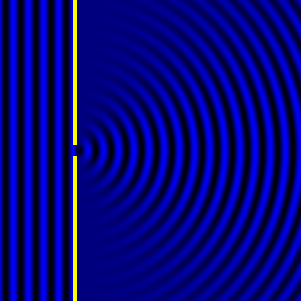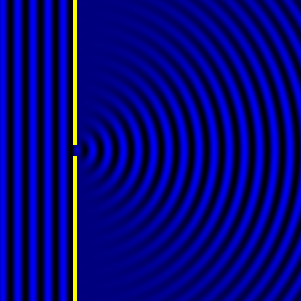
- Diffraction pattern caused by a slit of width equivalent to one wavelength [1].
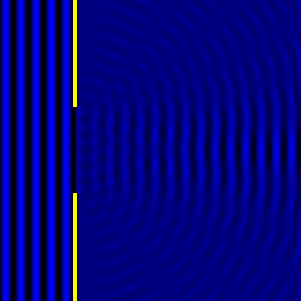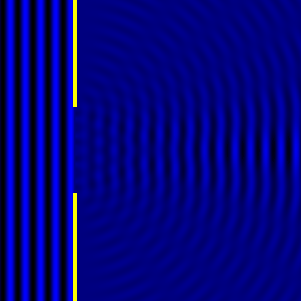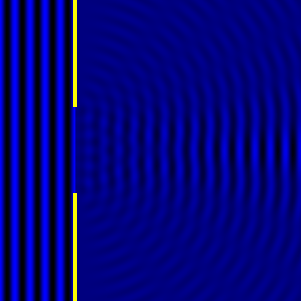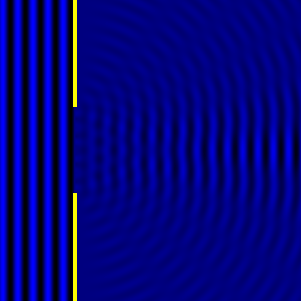
- Diffraction pattern caused by a slit of width equivalent to six wavelengths [1].
The above images show examples of patterns being formed in wavefronts by apertures in the path of otherwise uninterrupted waves. As can be seen by contrasting these images, the pattern created by a larger aperture is distinguishable from a smaller aperture's pattern. The particular distinctions can be described through mathematical formalisms. Thence, once the diffraction patterns have been created, it is possible to infer the shape and size of the objects that caused them, if we also know the wavelength of the light source and the distance from the sample at which the patterns were observed.
Co-Cultures: Bacteria and Algae can be Friends!
Go Together, Grow Together.
This section is still under development, but it will soon contain information regarding a co-culture comprising Chlamydomonas reinhardtii and Escherichia coli.
References
- [1]: Diffraction Formalism, Wikipedia, as on 4th September 2017 at http://en.wikipedia.org/wiki/Diffraction_formalism



