Jol-Fengzi (Talk | contribs) |
|||
| (136 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:SUSTech_Shenzhen/removeStyles}} | {{:Team:SUSTech_Shenzhen/removeStyles}} | ||
| − | {{:Team:SUSTech_Shenzhen/nav | + | {{:Team:SUSTech_Shenzhen/nav}}<!--------子页面导航栏------------------------------------> |
{{:Team:SUSTech_Shenzhen/themeCss}}<!--------整体布局,勿动-------------------------------> | {{:Team:SUSTech_Shenzhen/themeCss}}<!--------整体布局,勿动-------------------------------> | ||
<!------------以下空行应当保留-------------> | <!------------以下空行应当保留-------------> | ||
| + | <!--{{:Team:SUSTech_Shenzhen/templates/page-header-hardware|title=Hardware|subtitle=Create for wisdom of Life}}--> | ||
| + | <html> | ||
| + | <head> | ||
| + | <!--link href="../../assets/css/ie10-viewport-bug-workaround.css" rel="stylesheet"--> | ||
| + | <style type="text/css"> | ||
| + | :root #content > #right > .dose > .dosesingle, | ||
| + | :root #content > #center > .dose > .dosesingle | ||
| + | {display:none !important;}</style> | ||
| + | <!-- Custom styles for this template --> | ||
| + | <link href="https://2017.igem.org/wiki/index.php?title=Team:SUSTech_Shenzhen/themeCss/carousel-css&action=edit" rel="stylesheet"> | ||
| + | </head> | ||
| + | <!-- NAVBAR | ||
| + | ================================================== --> | ||
| + | <body style=""> | ||
| + | <div id="myCarousel" class="carousel slide" data-ride="carousel"> | ||
| + | <!-- Indicators --> | ||
| + | <ol class="carousel-indicators"> | ||
| + | <li data-target="#myCarousel" data-slide-to="0" class="active"></li> | ||
| + | <li data-target="#myCarousel" data-slide-to="1" class=""></li> | ||
| + | <li data-target="#myCarousel" data-slide-to="2" class=""></li> | ||
| + | <li data-target="#myCarousel" data-slide-to="3" class=""></li> | ||
| + | </ol> | ||
| + | <div class="carousel-inner" role="listbox"> | ||
| + | <div class="item active"> | ||
| + | <img class="first-slide" src="https://static.igem.org/mediawiki/2017/4/4f/T--SUSTech_Shenzhen--Microfluidics1overview.jpeg" alt="First slide" > | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption"> | ||
| + | <h1 style="font-size:54px">Microfluidics</h1> | ||
| + | <p style="font-size:24px">A function to research <i>C. elegans</i>'s neuron activity and behavioral response </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img class="second-slide" src="https://static.igem.org/mediawiki/2017/7/7c/T--SUSTech_Shenzhen--Microfluidics2overview.jpeg" alt="Second slide"> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption"> | ||
| + | <h1 style="font-size:54px">Pump and microscope </h1> | ||
| + | <p style="font-size:24px">A platform to use microfluidics</p> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img class="third-slide" src="https://static.igem.org/mediawiki/2017/5/52/T--SUSTech_Shenzhen--light1.jpeg" alt="Third slide"> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption"> | ||
| + | <h1 style="font-size:54px">Projector</h1> | ||
| + | <p style="font-size:24px">A custom light source to emit the blue light and red light </p> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img class="fourth-slide" src="https://static.igem.org/mediawiki/2017/2/25/T--SUSTech_Shenzhen--light2.jpeg" alt="Fourth slide"> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption"> | ||
| + | <h1 style="font-size:54px">Microscope and projector</h1> | ||
| + | <p style="font-size:24px"> Play with light in spatio-temporal</p> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <a class="left carousel-control" href="#myCarousel" role="button" data-slide="prev"> | ||
| + | <span class="glyphicon glyphicon-chevron-left" aria-hidden="true"></span> | ||
| + | <span class="sr-only">Previous</span> | ||
| + | </a> | ||
| + | <a class="right carousel-control" href="#myCarousel" role="button" data-slide="next"> | ||
| + | <span class="glyphicon glyphicon-chevron-right" aria-hidden="true"></span> | ||
| + | <span class="sr-only">Next</span> | ||
| + | </a> | ||
| + | </div><!-- /.carousel --> | ||
| + | <div class="container marketing"> | ||
| − | + | ||
| − | + | </div><!-- /.container --> | |
| − | + | ||
| − | <!-- | + | |
| − | + | ||
| − | + | </body></html> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | + | {{:Team:SUSTech_Shenzhen/main-content-begin}}<!---------内容的样式----------------> | |
| − | + | <!--------最上端的图片------------------------------------> | |
| − | + | <!--------往下直接写内容-----------------> | |
| − | + | ------------------------------------ | |
| − | + | = Microfluidics = | |
| − | + | ||
| − | + | In our experiment, we want to study the neural network activity and behavioral response of <i>Caenorhabditis elegans </i>under light stimuli of specific wavelengths. Thus, we need to design an accurate and user-friendly platform for studying the collective behavior of worms with high throughput as well as live neuron-level observations under natural conditions. | |
| − | </ | + | |
| − | + | ||
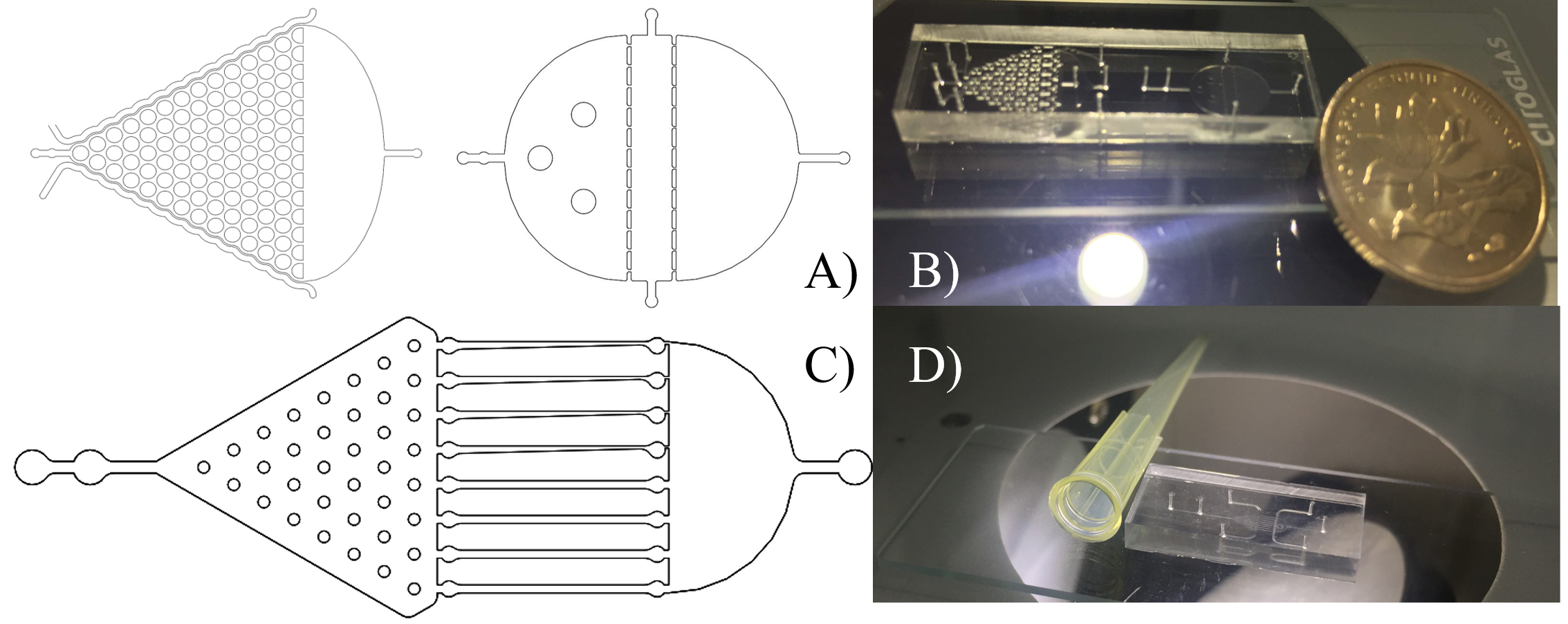
| + | To meet these design goals, we designed three microfluidic chips: the Selection Chip to select worms of appropriate sizes, the Gaussian Plate to monitor changes in the worm’s group behavior, and the Immobilization Plate to observe live neuron activities without anesthetization. (Fig.1) | ||
| − | |||
| − | = | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Microfluidics--fig1overview.png |width=1000px| caption=<B>Fig. 1 Three microfluidic systems in our experiment. A) </B>Design of the Selection Chip and the Gaussian Chip.<B> B) </B>The Selection Chip and the Gaussian Chip, fabricated on the same substrate <B>C)</B> Design of the Immobilization Chip.<B> D)</B> The Immobilization Chip.}} |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <html><a target="_black" href="https://2017.igem.org/Team:SUSTech_Shenzhen/Hardware/Microfluidics" class="btn btn-default"><i class="ion-arrow-right-c"></i> Detailed Microfluidics</a></html> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <html> | + | |
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
| − | </html> | + | |
| + | ------------------------------- | ||
| + | = Light Modulator = | ||
| + | Multiple devices of optics are designed and created for the various experiment requirements, such as, stimulate neuron of ''C. elegans'', train ''C. elegans'' and induce ''C. elegans'' move in a special direction. All devices attempts to modulate the spatio-temporal pattern in an '''elegant''' and '''effective''' way. We constructed projector light source and modulated mercury lamp. Just let us start to play with light. | ||
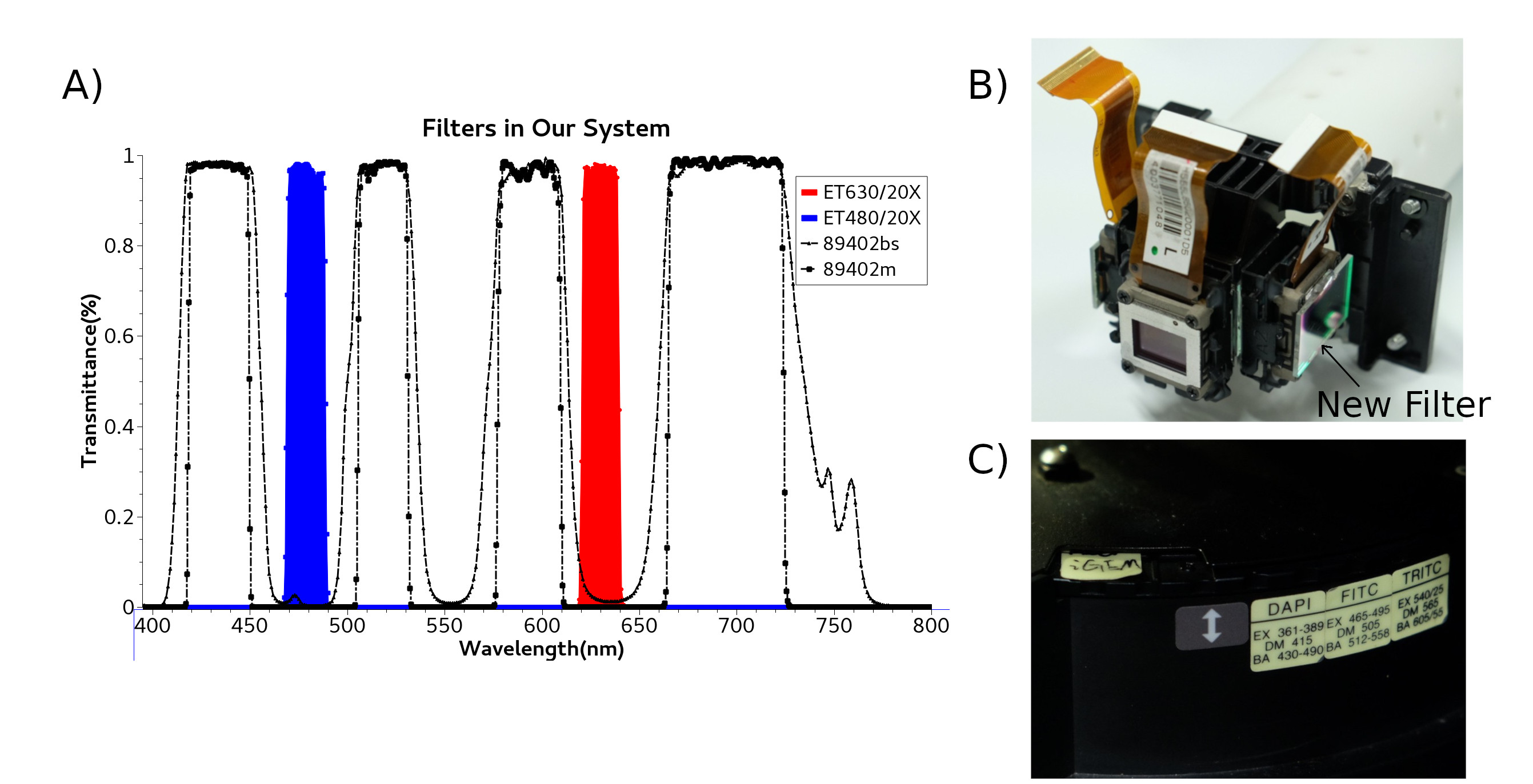
| + | We use 395nm light to activate calcium indicator protein GEM-GECO by Lumencor LED Illuminator. Then CoChR and Chrimson are activated by blue light and red light from LCD projector. LCD projector and LED Illuminator are merged by a double LH Adapter contained a semi-transparent mirror, which connect to microscope. So we can use these two light source in the same time. To achieve the aim, we add additional filter to purify the red light and blue light and replace new lens to adjust the focus distance(Fig. 2). | ||
| + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--SetofFilter.png| width=10000px | caption=Fig. 2 A) Spectra of filters and mirrors are used in our system. B) Filter ET630/20X is for Chrimson, and Filter ET480/20x is for CoChR. Both these are installed outside 3LCDs. C) Di-mirror and emission filter are installed inside microscope}} | ||
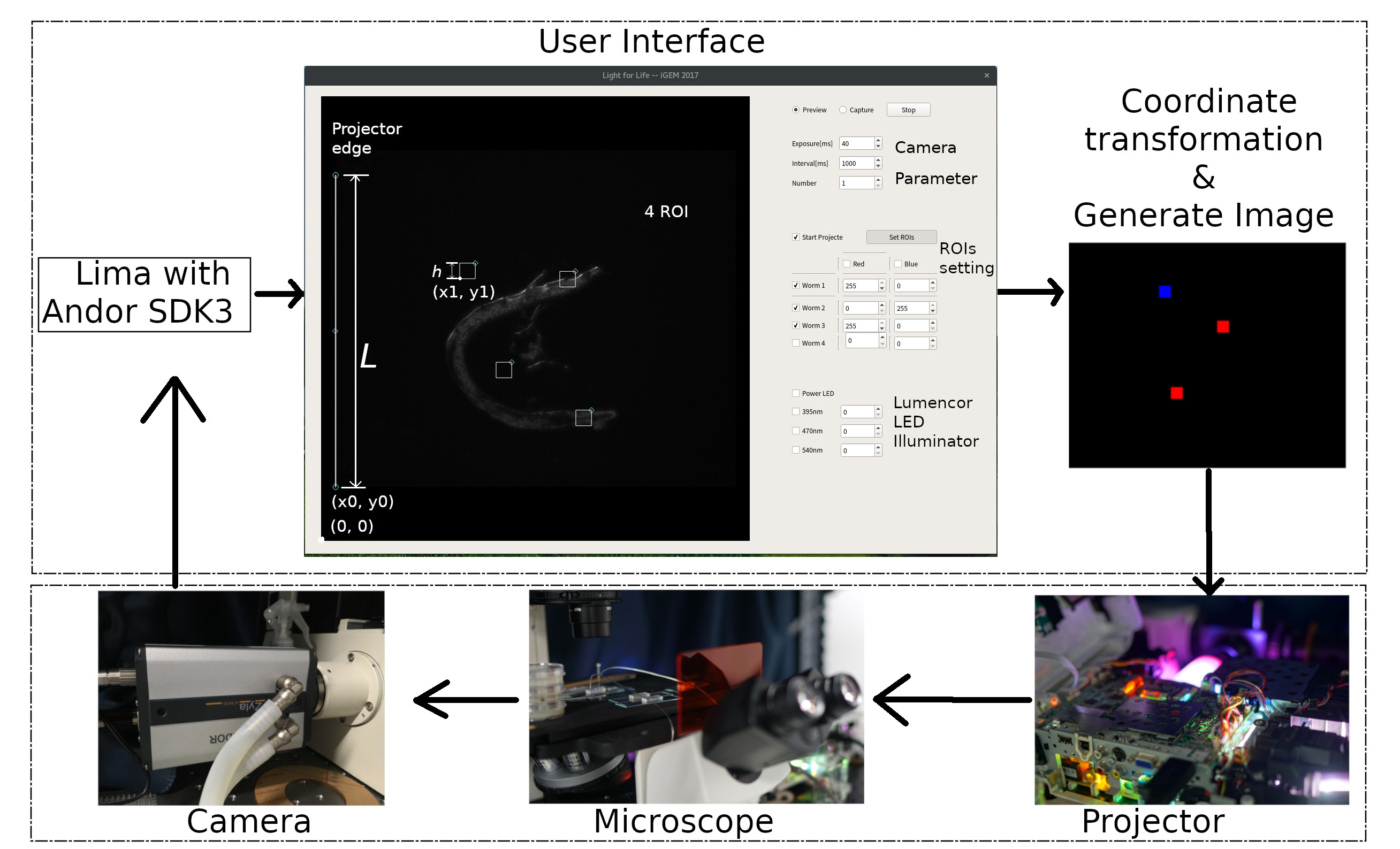
| − | + | We try develop a powerful and hackable open source software suite called ''ColorMapping'' to track and activate multi ''<i>C. elegans</i>'' or cell independently in one view(Fig. 3). User can modify multi color, intensity, time, locations of light alternately. ''ColorMapping'' can be found in [https://github.com/JiangXL/ColorMapping GitHub], which still is developing. | |
| − | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--LightModulator_Software.jpg |width=10003px | caption='''Fig. 3 The design's flowchart of ''ColorMapping'' '''. Image display in user interface from camera by Lima with Andor SDK3. User can track worm with some parameter. Then software generate image and project into microscope from projector. Finally, camera captures these pattern.}} | |
| − | + | ||
| − | |||
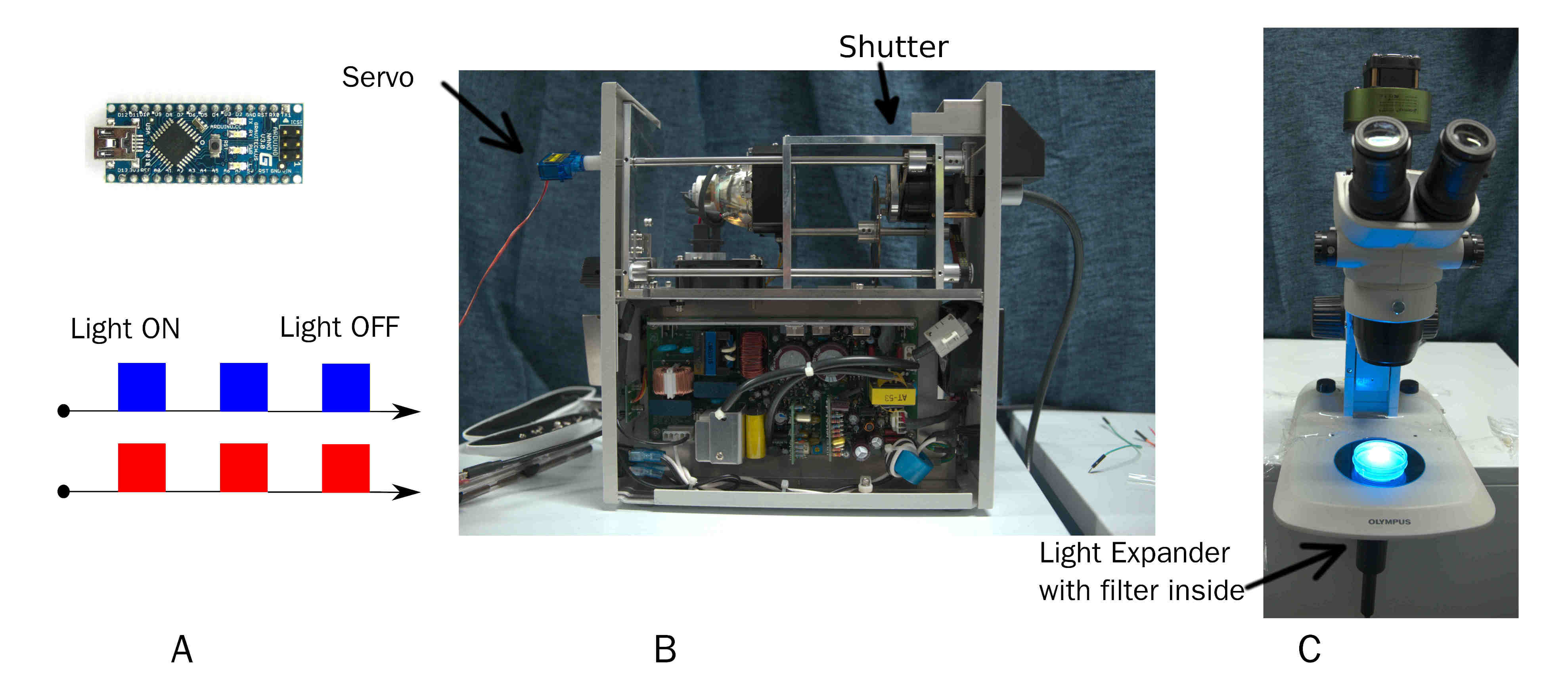
| + | For training ''C. elegans'', we develop a simple and effective device to output pulse of certain wavelength of light. On time and off time of pulse is custom by chip. Wavelength of light is changed by replacing filter inside beam expander.(Fig. 4) | ||
| + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--DSCF2143.jpg |width=1000px| caption='''Fig. 4 Overview of Arduino modulate Mercury lamp'''(A) Arduino transfer the pulse command to servo. (B)The interview of Mercury Lamp.The servo fix at the original bottom. (C) The beam expander connect to stereoscope}} | ||
| + | <html><a target="_black" href="https://2017.igem.org/Team:SUSTech_Shenzhen/Hardware/Light" class="btn btn-default"><i class="ion-arrow-right-c"></i> Detailed Light Modulator</a></html> | ||
| − | |||
Latest revision as of 16:51, 14 December 2017
Contents
Microfluidics
In our experiment, we want to study the neural network activity and behavioral response of Caenorhabditis elegans under light stimuli of specific wavelengths. Thus, we need to design an accurate and user-friendly platform for studying the collective behavior of worms with high throughput as well as live neuron-level observations under natural conditions.
To meet these design goals, we designed three microfluidic chips: the Selection Chip to select worms of appropriate sizes, the Gaussian Plate to monitor changes in the worm’s group behavior, and the Immobilization Plate to observe live neuron activities without anesthetization. (Fig.1)
Light Modulator
Multiple devices of optics are designed and created for the various experiment requirements, such as, stimulate neuron of C. elegans, train C. elegans and induce C. elegans move in a special direction. All devices attempts to modulate the spatio-temporal pattern in an elegant and effective way. We constructed projector light source and modulated mercury lamp. Just let us start to play with light.
We use 395nm light to activate calcium indicator protein GEM-GECO by Lumencor LED Illuminator. Then CoChR and Chrimson are activated by blue light and red light from LCD projector. LCD projector and LED Illuminator are merged by a double LH Adapter contained a semi-transparent mirror, which connect to microscope. So we can use these two light source in the same time. To achieve the aim, we add additional filter to purify the red light and blue light and replace new lens to adjust the focus distance(Fig. 2).
We try develop a powerful and hackable open source software suite called ColorMapping to track and activate multi C. elegans or cell independently in one view(Fig. 3). User can modify multi color, intensity, time, locations of light alternately. ColorMapping can be found in GitHub, which still is developing.
For training C. elegans, we develop a simple and effective device to output pulse of certain wavelength of light. On time and off time of pulse is custom by chip. Wavelength of light is changed by replacing filter inside beam expander.(Fig. 4)