Jol-Fengzi (Talk | contribs) |
|||
| Line 7: | Line 7: | ||
{{:Team:SUSTech_Shenzhen/main-content-begin}}<!---------内容的样式----------------> | {{:Team:SUSTech_Shenzhen/main-content-begin}}<!---------内容的样式----------------> | ||
<!--------往下直接写内容-----------------> | <!--------往下直接写内容-----------------> | ||
| − | |||
| − | The | + | ==Part:BBa_K2492002== |
| + | |||
| + | To indicate neuron AWA in <i>C.elegans</i>’(Caenorhabditis elegans) head, we chose mCherry. We found that the part BBa_K2005050 is mCherry submitted by iGEM16_Vanderbilt. However, their mCherry was not designed for <i>C.elegans</i>. | ||
| + | |||
| + | So, we got the sequence and improved it with <a href=" https://worm.mpi-cbg.de/codons/cgi-bin/optimize.py"> <i>C.elegans</i> codon adaptor</a> <ref>Redemann, S., Schloissnig, S., Ernst, S., Pozniakowsky, A., Ayloo, S., & Hyman, A. A., et al. (2011). Codon adaptation-based control of protein expression in <i>C.elegans</i>. Nature Methods, 8(3), 250.</ref>. Compared with original mCherry, new synthesized mCherry sequence is modified in two ways, three introns were added, and the codons were modified(Figure 1). | ||
| + | |||
| + | {{SUSTech_Image_Center_8 | filename=|width=1000px|caption=<B>Figure 1:Alignment between BBa_K2005050 and BBa_K2492002(optimized mCherry).</B>}} | ||
| + | |||
| + | The mCherry part we using now is optimized for <i>C.elegans</i>. We added the characterization to part page since the original part has no visual characterization neither on part page nor on their wiki. However, our mCherry is expressed in <i>C.elegans</i>. Plate reader and flow cytometer are not available for fluorescence measurement. | ||
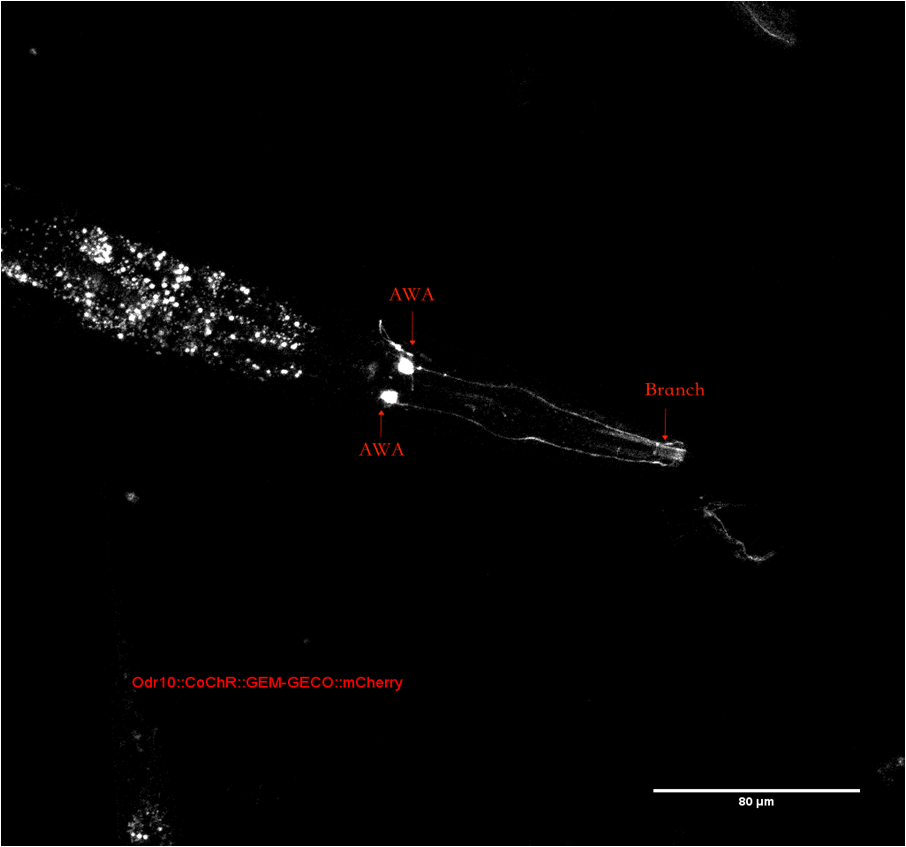
| + | To confirm the position of neuron AWA in <i>C.elegans</i>’ head. We observe <i>C.elegans</i> with a confocal microscope(Figure 2). The whole neuron with dendrite can be observed distinctly. The branch of dendrite near the mouse can be observed as described in <a href="http://www.wormatlas.org/neurons/Individual%20Neurons/AWAframeset.html">wormatlas</a> | ||
| + | |||
| + | {{SUSTech_Image_Center_8 | filename=T--SUSTech_Shenzhen--Parts-Improvement.png|width=1000px|caption=<B>Figure 2: The head of <i>C. elegans</i>, arrows point to AWA stoma and one of the branches</B>}} | ||
| + | |||
| + | == References == | ||
| + | <references /> | ||
| + | |||
| − | |||
{{:Team:SUSTech_Shenzhen/main-content-end}}<!---------内容的样式-----------------> | {{:Team:SUSTech_Shenzhen/main-content-end}}<!---------内容的样式-----------------> | ||
{{:Team:SUSTech_Shenzhen/wiki-footer}} | {{:Team:SUSTech_Shenzhen/wiki-footer}} | ||
{{:Team:SUSTech_Shenzhen/themeJs}}<!--------布局居中-------------> | {{:Team:SUSTech_Shenzhen/themeJs}}<!--------布局居中-------------> | ||
Revision as of 08:42, 31 October 2017
Improvement
Contribution
Contents
Part:BBa_K2492002
To indicate neuron AWA in C.elegans’(Caenorhabditis elegans) head, we chose mCherry. We found that the part BBa_K2005050 is mCherry submitted by iGEM16_Vanderbilt. However, their mCherry was not designed for C.elegans.
So, we got the sequence and improved it with <a href=" https://worm.mpi-cbg.de/codons/cgi-bin/optimize.py"> C.elegans codon adaptor</a> [1]. Compared with original mCherry, new synthesized mCherry sequence is modified in two ways, three introns were added, and the codons were modified(Figure 1).
The mCherry part we using now is optimized for C.elegans. We added the characterization to part page since the original part has no visual characterization neither on part page nor on their wiki. However, our mCherry is expressed in C.elegans. Plate reader and flow cytometer are not available for fluorescence measurement. To confirm the position of neuron AWA in C.elegans’ head. We observe C.elegans with a confocal microscope(Figure 2). The whole neuron with dendrite can be observed distinctly. The branch of dendrite near the mouse can be observed as described in <a href="http://www.wormatlas.org/neurons/Individual%20Neurons/AWAframeset.html">wormatlas</a>
References
- ↑ Redemann, S., Schloissnig, S., Ernst, S., Pozniakowsky, A., Ayloo, S., & Hyman, A. A., et al. (2011). Codon adaptation-based control of protein expression in C.elegans. Nature Methods, 8(3), 250.