Chemical Diffusion
Model
Contents
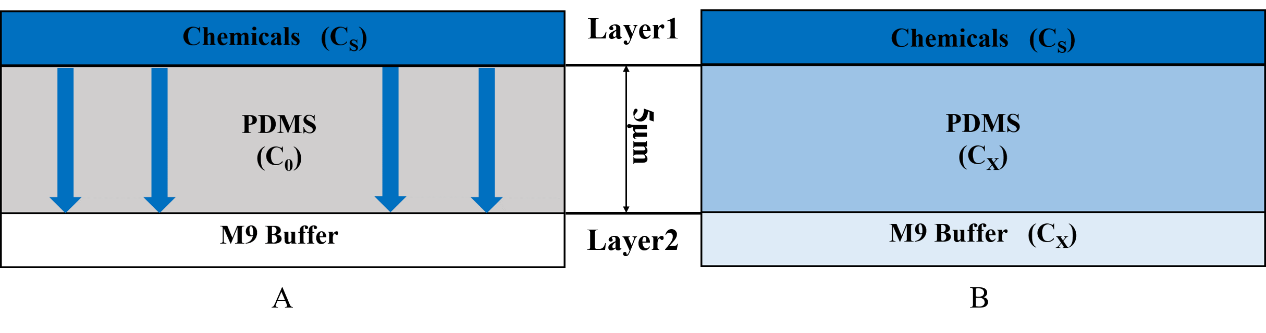
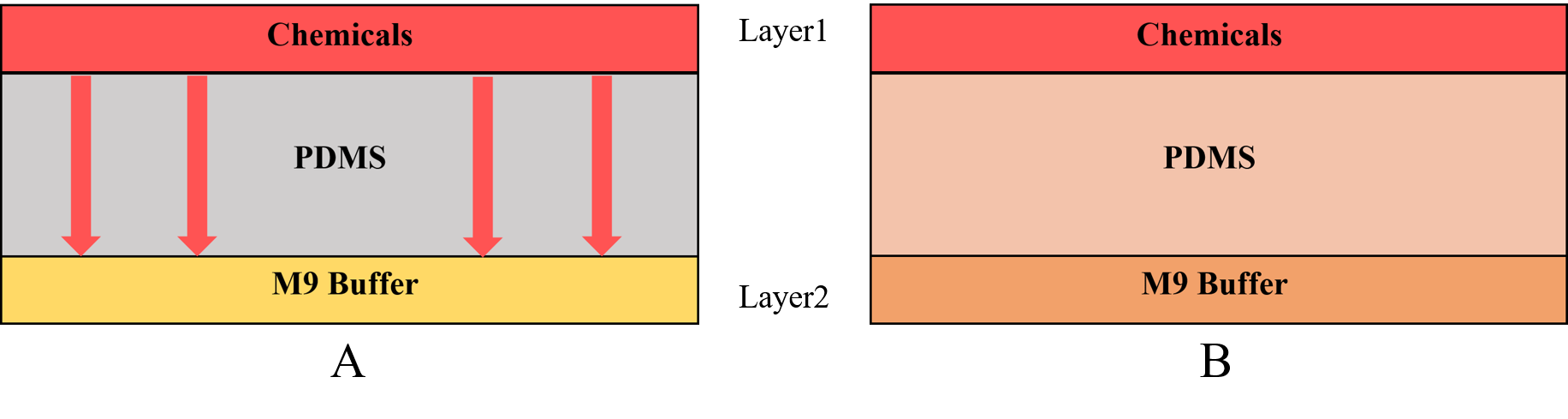
In our project, we are required to insert a new gene into the Caenorhabditis elegans’ (C. elegans') genome and express two channelrhodopsins in the olfactory receptor neuron pair. Because of the exogenous genes, we need a device to test if those exogenous genes will affect the olfactory receptor neuron pair. According to the Galton plate, we designed the Gaussian Plate[1] to test our worms (Fig.1 Gaussian Plate)
Using this chip we can know the C. elegans’ preference and repulsion through their distribution in the chip. This plate consists of two layers (Fig.1). We inject chemicals into the first layer to form a concentration gradient (Fig.2), then the chemicals with different concentration diffuse downward across the PDMS layer and reach the second layer. Then the worms can sense the chemicals.
In order to get the time when the worms in second layer can sense the chemicals we built the diffusion model to know the diffusion in PDMS. (Fig.3).
Diffusion
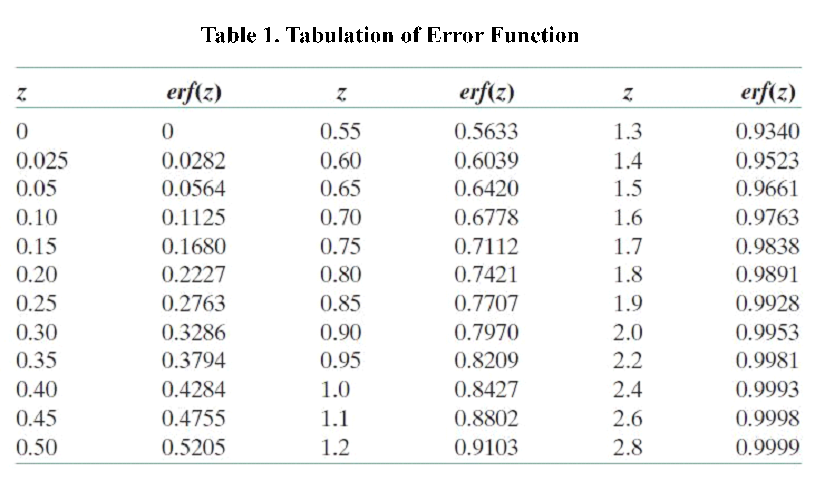
For nonsteady-state diffusion, there is an equation named Fick’s second law[2]: \frac{\partial C}{\partial t}=D \frac{\partial^2 C}{\partial x^2}
The constant of proportionality D is called the diffusion coefficient, which is expressed in square meters per second. Concentration C is plotted versus position (or distance) within the solid x and the time t.
For t=0, C=C0 at 0≤x≤∞;
For t>0, C=Cs (the constant surface concentration) at x=0;
C=C0 at x=∞. And then there is a general solution: \frac{C_x-C_0}{C_s-C_0}=1-erf(\frac{x}{2 \sqrt{Dt}}) Where Cx represents the concentration at distance x after time t.
\frac{x}{2 \sqrt{Dt}}=constant, so if this constant can be calculated, the equation of distance x and time t can also be gotten.
In our model, C0=0, so the equation is changed to \frac{C_x}{C_s}=1-erf(\frac{x}{2 \sqrt{Dt}}) Cx represents the concentration at distance x that C. elegans can feel. Cs is the concentration set by our own and it is a constant. x is 5mm (Fig.4).
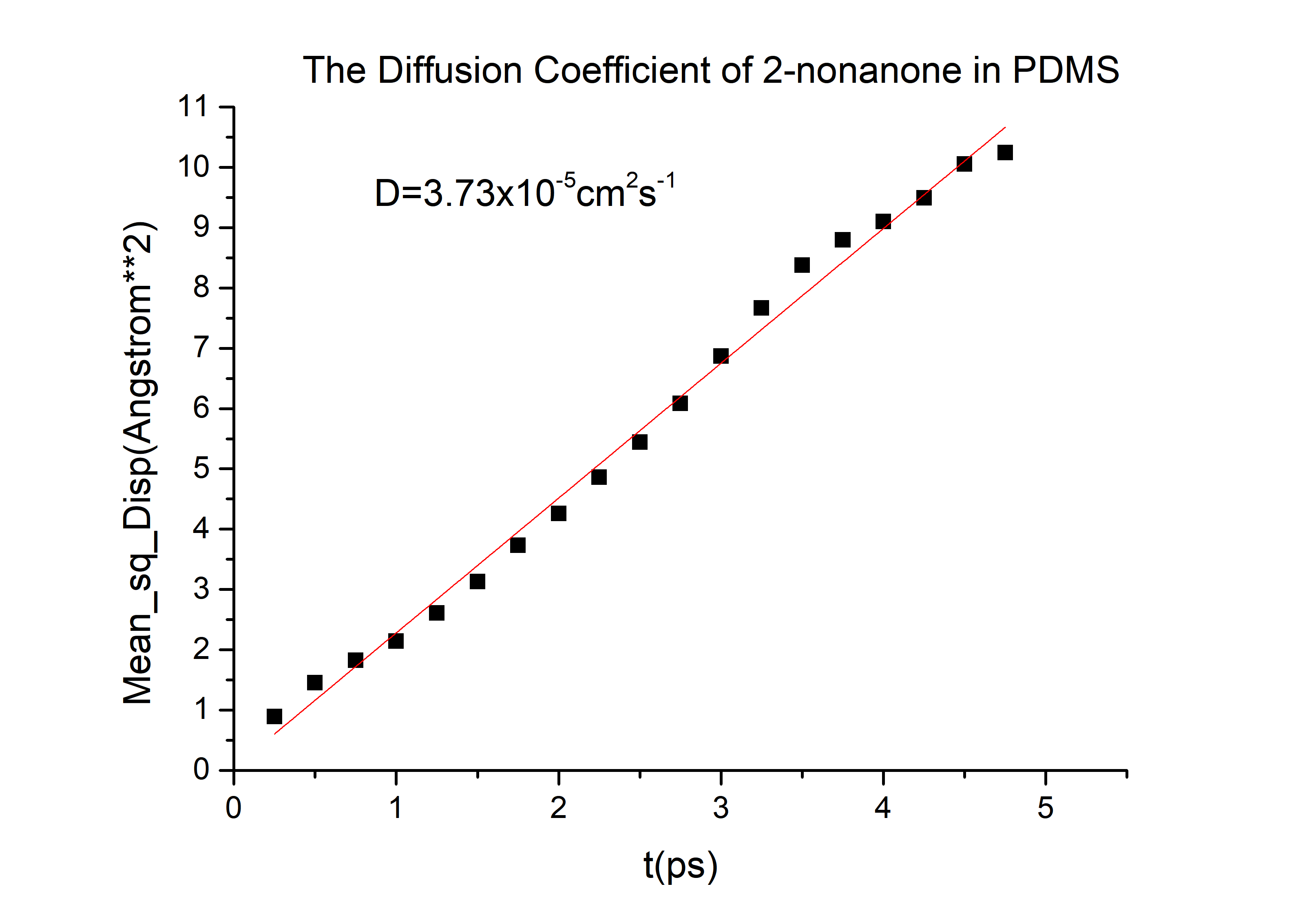
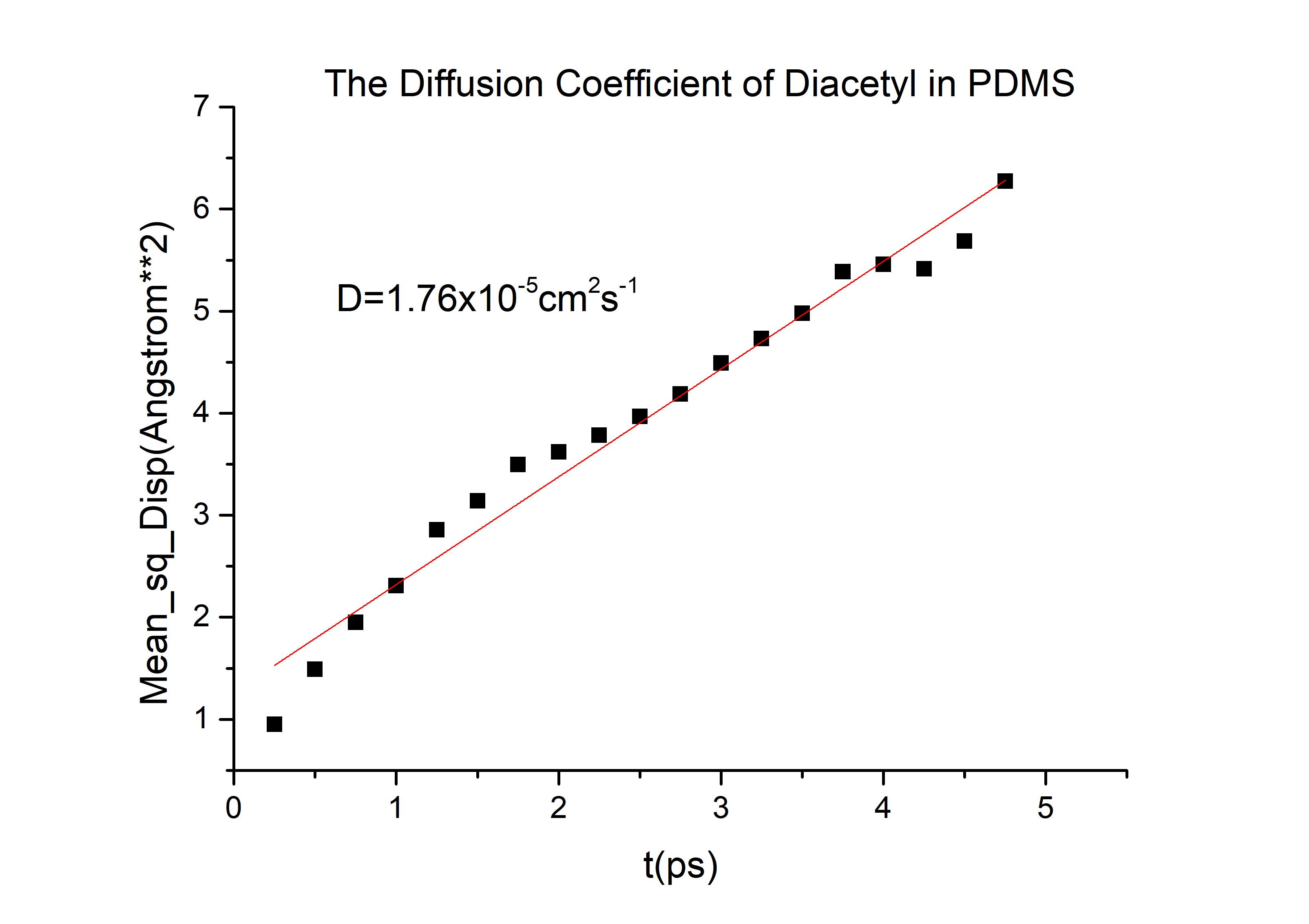
According to the equation we can get that the erf (\frac{x}{2 \sqrt{Dt}})=0.9965 and then referred to the Table 1.1 we can get that the \frac{x}{2 \sqrt{Dt}}=2.2, and D is the diffusion coefficient for 2-nonanone or diacetyl in PDMS. We need to calculate the D to get the time t according to the equation. We used the force-field method to get the D of those two chemicals (the concrete operational process is shown in 1.3).
D(2-nonanone) = 6.78*10-6cm2/s
D(diacetyl) = 6.78*10-6cm2/s
Finally, we can get the time t = 12 mins, on the other words, we cannot inject the C. elegans until the chemicals are injecting for 12 mins in layer 1(for diacetyl).
The Calculation of Diffusion Coefficient D
The diffusivity of a gas in an organic solvent, polymer, or zeolite can be calculated by running a molecular dynamics simulation and determining the mean square displacement of the gas in the material. This allows us to calculate the self-diffusivity coefficient of the gas and gives an insight into the overall diffusivity. As we are performing a molecular dynamics calculation, we can analyze the effect of temperature, pressure, density, and penetrant size and structure on diffusion. We used force-field method to calculate the D in PDMS by Material Studio (MS) a software for material calculation.[3][4]
Set up the initial structures
First, we set up the initial structure of the PDMS and the chemicals (Fig.5 and Fig.6).
Build an amorphous cell
Then, we put the two molecules in to an amorphous cell (Fig.7).
Relax the cell
When we generate an amorphous cell, the molecules may not be equally distributed throughout the cell, creating areas of vacuum. To correct this, we must perform a short energy minimization to optimize the cell. After the minimization, we should run a short molecular dynamics simulation to equilibrate the cell.
Run and analyze molecular dynamics
Export data and calculate the diffusivity
Then we can got the D of 2-nonanone and diacetyl:
D(2-nonanone) = 6.78*10-6cm2/s
D(diacetyl) = 6.78*10-6cm2/s
References
- ↑ Albrecht, D. R. and C. I. Bargmann (2011). “High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments.” Nature Methods 8(7): 599-605.
- ↑ Callister, W. D., & Rethwisch, D. G. (2004). Fundamentals of Materials Science and Engineering. John Wiley and Sons Ltd.
- ↑ S. G. Charati† and, & Stern, S. A. (1998). Diffusion of gases in silicone polymers: molecular dynamics simulations. Macromolecules, 31(16), 5529-5535.
- ↑ Hofmann, D., Fritz, L., Ulbrich, J., Schepers, C., & Böhning, M. (2000). Detailed‐atomistic molecular modeling of small molecule diffusion and solution processes in polymeric membrane materials. Macromolecular Theory & Simulations, 9(6), 293–327.